Genetic insights into human cortical organization and development through genome-wide analyses of 2,347 neuroimaging phenotypes
- PMID: 37592024
- PMCID: PMC10600728
- DOI: 10.1038/s41588-023-01475-y
Genetic insights into human cortical organization and development through genome-wide analyses of 2,347 neuroimaging phenotypes
Abstract
Our understanding of the genetics of the human cerebral cortex is limited both in terms of the diversity and the anatomical granularity of brain structural phenotypes. Here we conducted a genome-wide association meta-analysis of 13 structural and diffusion magnetic resonance imaging-derived cortical phenotypes, measured globally and at 180 bilaterally averaged regions in 36,663 individuals and identified 4,349 experiment-wide significant loci. These phenotypes include cortical thickness, surface area, gray matter volume, measures of folding, neurite density and water diffusion. We identified four genetic latent structures and causal relationships between surface area and some measures of cortical folding. These latent structures partly relate to different underlying gene expression trajectories during development and are enriched for different cell types. We also identified differential enrichment for neurodevelopmental and constrained genes and demonstrate that common genetic variants associated with cortical expansion are associated with cephalic disorders. Finally, we identified complex interphenotype and inter-regional genetic relationships among the 13 phenotypes, reflecting the developmental differences among them. Together, these analyses identify distinct genetic organizational principles of the cortex and their correlates with neurodevelopment.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures
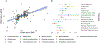
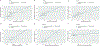
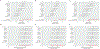


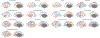





References
Publication types
MeSH terms
Grants and funding
- U01 DA051039/DA/NIDA NIH HHS/United States
- U01 DA041120/DA/NIDA NIH HHS/United States
- U01 DA051018/DA/NIDA NIH HHS/United States
- U01 DA041093/DA/NIDA NIH HHS/United States
- K08 MH120564/MH/NIMH NIH HHS/United States
- U24 DA041123/DA/NIDA NIH HHS/United States
- U01 DA051037/DA/NIDA NIH HHS/United States
- U01 DA051016/DA/NIDA NIH HHS/United States
- U01 DA041106/DA/NIDA NIH HHS/United States
- U01 DA041117/DA/NIDA NIH HHS/United States
- U01 DA041148/DA/NIDA NIH HHS/United States
- 214322/WT_/Wellcome Trust/United Kingdom
- R01 MH123922/MH/NIMH NIH HHS/United States
- U01 DA041174/DA/NIDA NIH HHS/United States
- U24 DA041147/DA/NIDA NIH HHS/United States
- T32 MH019112/MH/NIMH NIH HHS/United States
- U01 DA051038/DA/NIDA NIH HHS/United States
- R01 MH121521/MH/NIMH NIH HHS/United States
- U01 DA041134/DA/NIDA NIH HHS/United States
- U01 DA041022/DA/NIDA NIH HHS/United States
- U01 DA041156/DA/NIDA NIH HHS/United States
- U01 DA050987/DA/NIDA NIH HHS/United States
- U01 DA041025/DA/NIDA NIH HHS/United States
- U01 DA050989/DA/NIDA NIH HHS/United States
- U01 DA041089/DA/NIDA NIH HHS/United States
- U01 DA050988/DA/NIDA NIH HHS/United States
- U01 DA041028/DA/NIDA NIH HHS/United States
- U01 DA041048/DA/NIDA NIH HHS/United States
LinkOut - more resources
Full Text Sources

