Application of feature-based molecular networking and MassQL for the MS/MS fragmentation study of depsipeptides
- PMID: 37593127
- PMCID: PMC10427501
- DOI: 10.3389/fmolb.2023.1238475
Application of feature-based molecular networking and MassQL for the MS/MS fragmentation study of depsipeptides
Abstract
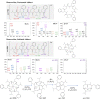
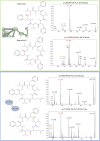
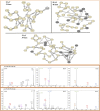
The Feature-based Molecular Networking (FBMN) is a well-known approach for mapping and identifying structures and analogues. However, in the absence of prior knowledge about the molecular class, assessing specific fragments and clusters requires time-consuming manual validation. This study demonstrates that combining FBMN and Mass Spec Query Language (MassQL) is an effective strategy for accelerating the decoding mass fragmentation pathways and identifying molecules with comparable fragmentation patterns, such as beauvericin and its analogues. To accomplish this objective, a spectral similarity network was built from ESI-MS/MS experiments of Fusarium oxysporum at various collision energies (CIDs) and paired with a MassQL search query for conserved beauvericin ions. FBMN analysis revealed that sodiated and protonated ions clustered differently, with sodiated adducts needing more collision energy and exhibiting a distinct fragmentation pattern. Based on this distinction, two sets of particular fragments were discovered for the identification of these hexadepsipeptides: ([M + H]+) m/z 134, 244, 262, and 362 and ([M + Na]+) m/z 266, 284 and 384. By using these fragments, MassQL accurately found other analogues of the same molecular class and annotated beauvericins that were not classified by FBMN alone. Furthermore, FBMN analysis of sodiated beauvericins at 70 eV revealed subclasses with distinct amino acid residues, allowing distinction between beauvericins (beauvericin and beauvericin D) and two previously unknown structural isomers with an unusual methionine sulfoxide residue. In summary, our integrated method revealed correlations between adduct types and fragmentation patterns, facilitated the detection of beauvericin clusters, including known and novel analogues, and allowed for the differentiation between structural isomers.
Keywords: MS/MS fragmentation; MassQL; PCA; beauvericin; feature-based molecular networking.
Copyright © 2023 Selegato, Zanatta, Pilon, Veloso and Castro-Gamboa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

