Structural aspects of enzymes involved in prokaryotic Gram-positive heme biosynthesis
- PMID: 37593721
- PMCID: PMC10427985
- DOI: 10.1016/j.csbj.2023.07.024
Structural aspects of enzymes involved in prokaryotic Gram-positive heme biosynthesis
Abstract
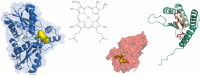
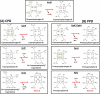
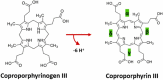
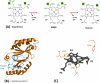
The coproporphyrin dependent heme biosynthesis pathway is almost exclusively utilized by Gram-positive bacteria. This fact makes it a worthwhile topic for basic research, since a fundamental understanding of a metabolic pathway is necessary to translate the focus towards medical biotechnology, which is very relevant in this specific case, considering the need for new antibiotic targets to counteract the pathogenicity of Gram-positive superbugs. Over the years a lot of structural data on the set of enzymes acting in Gram-positive heme biosynthesis has accumulated in the Protein Database (www.pdb.org). One major challenge is to filter and analyze all available structural information in sufficient detail in order to be helpful and to draw conclusions. Here we pursued to give a holistic overview of structural information on enzymes involved in the coproporphyrin dependent heme biosynthesis pathway. There are many aspects to be extracted from experimentally determined structures regarding the reaction mechanisms, where the smallest variation of the position of an amino acid residue might be important, but also on a larger level regarding protein-protein interactions, where the focus has to be on surface characteristics and subunit (secondary) structural elements and oligomerization. This review delivers a status quo, highlights still missing information, and formulates future research endeavors in order to better understand prokaryotic heme biosynthesis.
Keywords: Coproheme decarboxylase; Coproporphyrin ferrochelatase; Coproporphyrinogen oxidase; Frataxin; Molecular enzymology; Structure determination; Uroporphyrinogen decarboxylase.
© 2023 The Authors.
Conflict of interest statement
The authors of the submitted manuscript “Structural aspects of enzymes involved in prokaryotic Gram-positive heme biosynthesis” declare no conflict of interests.
Figures






Similar articles
-
Regulation of heme biosynthesis via the coproporphyrin dependent pathway in bacteria.Front Microbiol. 2024 Mar 21;15:1345389. doi: 10.3389/fmicb.2024.1345389. eCollection 2024. Front Microbiol. 2024. PMID: 38577681 Free PMC article. Review.
-
Insights into the flexibility of the domain-linking loop in actinobacterial coproheme decarboxylase through structures and molecular dynamics simulations.Protein Sci. 2025 Feb;34(2):e70027. doi: 10.1002/pro.70027. Protein Sci. 2025. PMID: 39865384 Free PMC article.
-
Crystal structures and calorimetry reveal catalytically relevant binding mode of coproporphyrin and coproheme in coproporphyrin ferrochelatase.FEBS J. 2020 Jul;287(13):2779-2796. doi: 10.1111/febs.15164. Epub 2019 Dec 19. FEBS J. 2020. PMID: 31794133 Free PMC article.
-
Prokaryotic Heme Biosynthesis: Multiple Pathways to a Common Essential Product.Microbiol Mol Biol Rev. 2017 Jan 25;81(1):e00048-16. doi: 10.1128/MMBR.00048-16. Print 2017 Mar. Microbiol Mol Biol Rev. 2017. PMID: 28123057 Free PMC article. Review.
-
Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin.Proc Natl Acad Sci U S A. 2015 Feb 17;112(7):2210-5. doi: 10.1073/pnas.1416285112. Epub 2015 Feb 2. Proc Natl Acad Sci U S A. 2015. PMID: 25646457 Free PMC article.
Cited by
-
Regulation of heme biosynthesis via the coproporphyrin dependent pathway in bacteria.Front Microbiol. 2024 Mar 21;15:1345389. doi: 10.3389/fmicb.2024.1345389. eCollection 2024. Front Microbiol. 2024. PMID: 38577681 Free PMC article. Review.
-
Insights into the flexibility of the domain-linking loop in actinobacterial coproheme decarboxylase through structures and molecular dynamics simulations.Protein Sci. 2025 Feb;34(2):e70027. doi: 10.1002/pro.70027. Protein Sci. 2025. PMID: 39865384 Free PMC article.
References
-
- Lobo S.A.L., Scott A., Videira M.A.M., Winpenny D., Gardner M., Palmer M.J., et al. Staphylococcus aureus Haem Biosynthesis: characterisation of the enzymes involved in final steps of the pathway: S. Aureus Haem Biosynthesis: from uroporphyrinogen III to haem. Mol Microbiol. 2015;97(3):472–487. doi: 10.1111/mmi.13041. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
