Mirikizumab-Induced Transcriptome Changes in Ulcerative Colitis Patient Biopsies at Week 12 Are Maintained Through Week 52
- PMID: 37594044
- PMCID: PMC10684203
- DOI: 10.14309/ctg.0000000000000630
Mirikizumab-Induced Transcriptome Changes in Ulcerative Colitis Patient Biopsies at Week 12 Are Maintained Through Week 52
Abstract
Introduction: Mirikizumab, an anti-interleukin-23p19 monoclonal antibody, demonstrated efficacy in phase 2 and 3 randomized clinical trials of patients with moderate-to-severe ulcerative colitis (UC). Previous results have shown that 12 weeks of mirikizumab treatment downregulated transcripts associated with UC disease activity and tumor necrosis factor inhibitor resistance. We assessed week-52 gene expression from week-12 responders receiving mirikizumab or placebo.
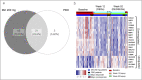
Methods: In the phase 2 AMAC study (NCT02589665), mirikizumab-treated patients achieving week-12 clinical response were rerandomized to mirikizumab 200 mg subcutaneous every 4 or 12 weeks through week 52 (N = 31). Week-12 placebo responders continued placebo through week 52 (N = 7). The limma R package clustered transcript changes in colonic mucosa biopsies from baseline to week 12 into differentially expressed genes (DEGs). Among DEGs, similarly expressed genes (DEGSEGs) maintaining week-12 expression through week 52 were identified.
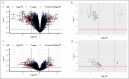
Results: Of 89 DEGSEGs, 63 (70.8%) were present only in mirikizumab induction responders, 5 (5.6%) in placebo responders, and 21 (23.6%) in both. Week-12 magnitudes and week-52 consistency of transcript changes were greater in mirikizumab than in placebo responders (log2FC > 1). DEGSEG clusters (from 84 DEGSEGs identified in mirikizumab and mirikizumab/placebo responders) correlated to modified Mayo score (26/84 with Pearson correlation coefficient [PCC] >0.5) and Robarts Histopathology Index (55/84 with PCC >0.5), sustained through week 52.
Discussion: Mirikizumab responders had broader, more sustained transcriptional changes of greater magnitudes at week 52 vs placebo. Mirikizumab responder DEGSEGs suggest a distinct molecular healing pathway associated with mirikizumab interleukin-23 inhibition. The cluster's correlation with disease activity illustrates relationships between clinical, endoscopic, and molecular healing in UC.
Copyright © 2023 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of The American College of Gastroenterology.
Conflict of interest statement
Figures




References
-
- Ordas I, Eckmann L, Talamini M, et al. Ulcerative colitis. Lancet 2012;380(9853):1606–19. - PubMed
-
- Peyrin-Biroulet L, Sandborn W, Sands BE, et al. Selecting therapeutic targets in inflammatory bowel disease (STRIDE): Determining therapeutic goals for treat-to-target. Am J Gastroenterol 2015;110(9):1324–38. - PubMed
-
- Turner D, Ricciuto A, Lewis A, et al. STRIDE-II: An update on the selecting therapeutic targets in inflammatory bowel disease (STRIDE) initiative of the international organization for the study of IBD (IOIBD): Determining therapeutic goals for treat-to-target strategies in IBD. Gastroenterology 2021;160(5):1570–83. - PubMed
-
- Moschen AR, Tilg H, Raine T. IL-12, IL-23 and IL-17 in IBD: Immunobiology and therapeutic targeting. Nat Rev Gastroenterol Hepatol 2019;16(3):185–96. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

