FOXA1 O-GlcNAcylation-mediated transcriptional switch governs metastasis capacity in breast cancer
- PMID: 37595040
- PMCID: PMC10438466
- DOI: 10.1126/sciadv.adg7112
FOXA1 O-GlcNAcylation-mediated transcriptional switch governs metastasis capacity in breast cancer
Abstract
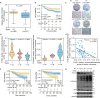
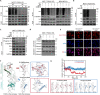
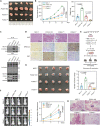
FOXA1, a transcription factor involved in epigenetic reprogramming, is crucial for breast cancer progression. However, the mechanisms by which FOXA1 achieves its oncogenic functions remain elusive. Here, we demonstrate that the O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) of FOXA1 promotes breast cancer metastasis by orchestrating the transcription of numerous metastasis regulators. O-GlcNAcylation at Thr432, Ser441, and Ser443 regulates the stability of FOXA1 and promotes its assembly with chromatin. O-GlcNAcylation shapes the FOXA1 interactome, especially triggering the recruitment of the transcriptional repressor methyl-CpG binding protein 2 and consequently stimulating FOXA1 chromatin-binding sites to switch to chromatin loci of adhesion-related genes, including EPB41L3 and COL9A2. Site-specific depletion of O-GlcNAcylation on FOXA1 affects the expression of various downstream genes and thus inhibits breast cancer proliferation and metastasis both in vitro and in vivo. Our data establish the importance of aberrant FOXA1 O-GlcNAcylation in breast cancer progression and indicate that targeting O-GlcNAcylation is a therapeutic strategy for metastatic breast cancer.
Figures






References
-
- K. M. Jozwik, J. S. Carroll, Pioneer factors in hormone-dependent cancers. Nat. Rev. Cancer 12, 381–385 (2012). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

