Two disulfide-reducing pathways are required for the maturation of plastid c-type cytochromes in Chlamydomonas reinhardtii
- PMID: 37595062
- PMCID: PMC10550313
- DOI: 10.1093/genetics/iyad155
Two disulfide-reducing pathways are required for the maturation of plastid c-type cytochromes in Chlamydomonas reinhardtii
Abstract
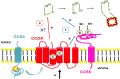
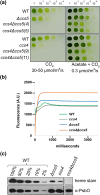
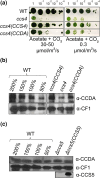
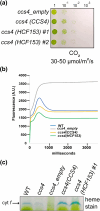
In plastids, conversion of light energy into ATP relies on cytochrome f, a key electron carrier with a heme covalently attached to a CXXCH motif. Covalent heme attachment requires reduction of the disulfide-bonded CXXCH by CCS5 and CCS4. CCS5 receives electrons from the oxidoreductase CCDA, while CCS4 is a protein of unknown function. In Chlamydomonas reinhardtii, loss of CCS4 or CCS5 yields a partial cytochrome f assembly defect. Here, we report that the ccs4ccs5 double mutant displays a synthetic photosynthetic defect characterized by a complete loss of holocytochrome f assembly. This defect is chemically corrected by reducing agents, confirming the placement of CCS4 and CCS5 in a reducing pathway. CCS4-like proteins occur in the green lineage, and we show that HCF153, a distant ortholog from Arabidopsis thaliana, can substitute for Chlamydomonas CCS4. Dominant suppressor mutations mapping to the CCS4 gene were identified in photosynthetic revertants of the ccs4ccs5 mutants. The suppressor mutations yield changes in the stroma-facing domain of CCS4 that restore holocytochrome f assembly above the residual levels detected in ccs5. Because the CCDA protein accumulation is decreased specifically in the ccs4 mutant, we hypothesize the suppressor mutations enhance the supply of reducing power through CCDA in the absence of CCS5. We discuss the operation of a CCS5-dependent and a CCS5-independent pathway controlling the redox status of the heme-binding cysteines of apocytochrome f.
Keywords: Chlamydomonas reinhardtii; Plant Genetics and Genomics; cytochromes c; energy-transducing membranes; photosynthesis; thiol–disulfide chemistry.
© The Author(s) 2023. Published by Oxford University Press on behalf of The Genetics Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

