E4 ubiquitin ligase promotes mitofusin turnover and mitochondrial stress response
- PMID: 37595558
- PMCID: PMC10434984
- DOI: 10.1016/j.molcel.2023.07.021
E4 ubiquitin ligase promotes mitofusin turnover and mitochondrial stress response
Abstract
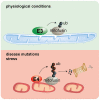
Ubiquitin-dependent control of mitochondrial dynamics is important for protein quality and neuronal integrity. Mitofusins, mitochondrial fusion factors, can integrate cellular stress through their ubiquitylation, which is carried out by multiple E3 enzymes in response to many different stimuli. However, the molecular mechanisms that enable coordinated responses are largely unknown. Here we show that yeast Ufd2, a conserved ubiquitin chain-elongating E4 enzyme, is required for mitochondrial shape adjustments. Under various stresses, Ufd2 translocates to mitochondria and triggers mitofusin ubiquitylation. This elongates ubiquitin chains on mitofusin and promotes its proteasomal degradation, leading to mitochondrial fragmentation. Ufd2 and its human homologue UBE4B also target mitofusin mutants associated with Charcot-Marie-Tooth disease, a hereditary sensory and motor neuropathy characterized by progressive loss of the peripheral nerves. This underscores the pathophysiological importance of E4-mediated ubiquitylation in neurodegeneration. In summary, we identify E4-dependent mitochondrial stress adaptation by linking various metabolic processes to mitochondrial fusion and fission dynamics.
Keywords: CMT2A; Cdc48/p97; E4; Fzo1; MFN2; UBE4B; Ufd2; fusion; mitochondria; mitofusin; stress; ubiquitin.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.B. received honorary for lectures and advisory boards from AbbVie, Amgen, AstraZeneca, Bayer, BMS, Boehringer-Ingelheim, Illumina, Janssen, Lilly, Merck-Serono, MSD, Novartis, Qiagen, Pfizer, Roche, Sanofi, and Targos MP Inc. R.B. is a co-founder and co-owner of Gnothis Inc (Stockholm SE) and Timer Therapeutics Inc (Freiburg and Fulda DE).
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

