FAM120A couples SREBP-dependent transcription and splicing of lipogenesis enzymes downstream of mTORC1
- PMID: 37595559
- PMCID: PMC10494788
- DOI: 10.1016/j.molcel.2023.07.017
FAM120A couples SREBP-dependent transcription and splicing of lipogenesis enzymes downstream of mTORC1
Abstract
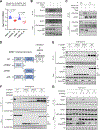
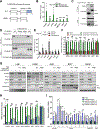
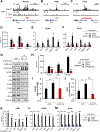
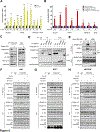
The mechanistic target of rapamycin complex 1 (mTORC1) is a master regulator of cell growth that stimulates macromolecule synthesis through transcription, RNA processing, and post-translational modification of metabolic enzymes. However, the mechanisms of how mTORC1 orchestrates multiple steps of gene expression programs remain unclear. Here, we identify family with sequence similarity 120A (FAM120A) as a transcription co-activator that couples transcription and splicing of de novo lipid synthesis enzymes downstream of mTORC1-serine/arginine-rich protein kinase 2 (SRPK2) signaling. The mTORC1-activated SRPK2 phosphorylates splicing factor serine/arginine-rich splicing factor 1 (SRSF1), enhancing its binding to FAM120A. FAM120A directly interacts with a lipogenic transcription factor SREBP1 at active promoters, thereby bridging the newly transcribed lipogenic genes from RNA polymerase II to the SRSF1 and U1-70K-containing RNA-splicing machinery. This mTORC1-regulated, multi-protein complex promotes efficient splicing and stability of lipogenic transcripts, resulting in fatty acid synthesis and cancer cell proliferation. These results elucidate FAM120A as a critical transcription co-factor that connects mTORC1-dependent gene regulation programs for anabolic cell growth.
Keywords: FAM120A; RNA splicing; RNA stability; SREBP; SRPK2; SRSF1; lipid metabolism; mTOR signaling.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.B. is an advisory board member for Molecular Cell.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

