Genome-wide analysis and characterization of the LRR-RLK gene family provides insights into anthracnose resistance in common bean
- PMID: 37596307
- PMCID: PMC10439169
- DOI: 10.1038/s41598-023-40054-3
Genome-wide analysis and characterization of the LRR-RLK gene family provides insights into anthracnose resistance in common bean
Abstract
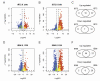
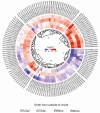
Anthracnose, caused by the hemibiotrophic fungus Colletotrichum lindemuthianum, is a damaging disease of common beans that can drastically reduce crop yield. The most effective strategy to manage anthracnose is the use of resistant cultivars. There are many resistance loci that have been identified, mapped and associated with markers in common bean chromosomes. The Leucine-rich repeat kinase receptor protein (LRR-RLK) family is a diverse group of transmembrane receptors, which potentially recognizes pathogen-associated molecular patterns and activates an immune response. In this study, we performed in silico analyses to identify, classify, and characterize common bean LRR-RLKs, also evaluating their expression profile in response to the infection by C. lindemuthianum. By analyzing the entire genome of Phaseolus vulgaris, we could identify and classify 230 LRR-RLKs into 15 different subfamilies. The analyses of gene structures, conserved domains and motifs suggest that LRR-RLKs from the same subfamily are consistent in their exon/intron organization and composition. LRR-RLK genes were found along the 11 chromosomes of the species, including regions of proximity with anthracnose resistance markers. By investigating the duplication events within the LRR-RLK family, we associated the importance of such a family with an expansion resulting from a strong stabilizing selection. Promoter analysis was also performed, highlighting cis-elements associated with the plant response to biotic stress. With regard to the expression pattern of LRR-RLKs in response to the infection by C. lindemuthianum, we could point out several differentially expressed genes in this subfamily, which were associated to specific molecular patterns of LRR-RLKs. Our work provides a broad analysis of the LRR-RLK family in P. vulgaris, allowing an in-depth structural and functional characterization of genes and proteins of this family. From specific expression patterns related to anthracnose response, we could infer a direct participation of RLK-LRR genes in the mechanisms of resistance to anthracnose, highlighting important subfamilies for further investigations.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- De Ron, A. M. et al. History of the common bean crop: its evolution beyond its areas of origin and domestication. Arbor. 192, 779:a317 (2016).
-
- Degu T, Yaregal W, Gudisa T. Status of common bean (Phaseolus vulgaris L.) diseases in metekel zone, North West Ethiopia. J. Plant Pathol. Microbiol. 2020;11:494.
-
- Carneiro, J. D. S., Paula Júnior, T. D. & Borém, A. Feij ao: Do plantio à colheita. UFV, Viçosa, 384p (2015).
-
- Padder B, Sharma P, Awale H, Kelly J. Colletotrichum lindemuthianum, the causal agent of bean anthracnose. J. Plant Pathol. 2017;99:2.
-
- Ferreira JJ, Campa A, Kelly JD. Organization of genes conferring resistance to anthracnose in common bean. Transl. Genomics Crop Breed. Biotic Stress. 2013;1:151–181.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

