Bacterial and archaeal community distributions and cosmopolitanism across physicochemically diverse hot springs
- PMID: 37596308
- PMCID: PMC10439147
- DOI: 10.1038/s43705-023-00291-z
Bacterial and archaeal community distributions and cosmopolitanism across physicochemically diverse hot springs
Abstract
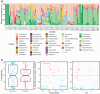
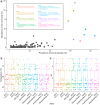
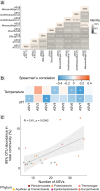
Terrestrial hot springs harbor diverse microbial communities whose compositions are shaped by the wide-ranging physico-chemistries of individual springs. The effect of enormous physico-chemical differences on bacterial and archaeal distributions and population structures is little understood. We therefore analysed the prevalence and relative abundance of bacteria and archaea in the sediments (n = 76) of hot spring features, in the Taupō Volcanic Zone (New Zealand), spanning large differences in major anion water chemistry, pH (2.0-7.5), and temperature (17.5-92.9 °C). Community composition, based on 16S rRNA amplicon sequence variants (ASVs) was strongly influenced by both temperature and pH. However, certain lineages characterized diverse hot springs. At the domain level, bacteria and archaea shared broadly equivalent community abundances across physico-chemically diverse springs, despite slightly lower bacteria-to-archaea ratios and microbial 16S rRNA gene concentrations at higher temperatures. Communities were almost exclusively dominated by Proteobacteria, Euryarchaeota or Crenarchaeota. Eight archaeal and bacterial ASVs from Thermoplasmatales, Desulfurellaceae, Mesoaciditogaceae and Acidithiobacillaceae were unusually prevalent (present in 57.9-84.2% of samples) and abundant (1.7-12.0% sample relative abundance), and together comprised 44% of overall community abundance. Metagenomic analyses generated multiple populations associated with dominant ASVs, and showed characteristic traits of each lineage for sulfur, nitrogen and hydrogen metabolism. Differences in metabolic gene composition and genome-specific metabolism delineated populations from relatives. Genome coverage calculations showed that populations associated with each lineage were distributed across a physicochemically broad range of hot springs. Results imply that certain bacterial and archaeal lineages harbor different population structures and metabolic potentials for colonizing diverse hot spring environments.
© 2023. ISME Publications B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Kent AD, Yannarell AC, Rusak JA, Triplett EW, McMahon KD. Synchrony in aquatic microbial community dynamics. ISME J. 2007;1:38–47. - PubMed
-
- Bodvarsson G. Physical characteristics of natural heat resources in Iceland. Joekull. 1961;11:29–38.
-
- Brock TD. Bimodal distribution of pH Values of thermal springs of the world. GSA Bull. 1971;82:1393–4.
-
- Jones B, Renaut RW, Rosen MR. Microbial construction of siliceous stalactites at geysers and hot springs: examples from the Whakarewarewa Geothermal Area, North Island, New Zealand. PALAIOS. 2001;16:73–94.

