Development of a novel glycoengineering platform for the rapid production of conjugate vaccines
- PMID: 37596672
- PMCID: PMC10436394
- DOI: 10.1186/s12934-023-02125-y
Development of a novel glycoengineering platform for the rapid production of conjugate vaccines
Abstract
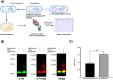
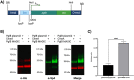
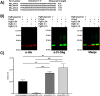
Conjugate vaccines produced either by chemical or biologically conjugation have been demonstrated to be safe and efficacious in protection against several deadly bacterial diseases. However, conjugate vaccine assembly and production have several shortcomings which hinders their wider availability. Here, we developed a tool, Mobile-element Assisted Glycoconjugation by Insertion on Chromosome, MAGIC, a novel biotechnological platform that overcomes the limitations of the current conjugate vaccine design method(s). As a model, we focused our design on a leading bioconjugation method using N-oligosaccharyltransferase (OTase), PglB. The installation of MAGIC led to at least twofold increase in glycoconjugate yield via MAGIC when compared to conventional N-OTase based bioconjugation method(s). Then, we improved MAGIC to (a) allow rapid installation of glycoengineering component(s), (b) omit the usage of antibiotics, (c) reduce the dependence on protein induction agents. Furthermore, we show the modularity of the MAGIC platform in performing glycoengineering in bacterial species that are less genetically tractable than the commonly used Escherichia coli. The MAGIC system promises a rapid, robust and versatile method to develop vaccines against serious bacterial pathogens. We anticipate the utility of the MAGIC platform could enhance vaccines production due to its compatibility with virtually any bioconjugation method, thus expanding vaccine biopreparedness toolbox.
Keywords: Bacterial vaccines; Bioconjugation; Conjugate vaccines; Glycoengineering.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
Brendan Wren and Jon Cuccui are founders of ArkVax Limited, a company that has an exclusive licence to the MAGIC technology (patent number 20150344928). All other authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

