Transposable elements as essential elements in the control of gene expression
- PMID: 37596675
- PMCID: PMC10439571
- DOI: 10.1186/s13100-023-00297-3
Transposable elements as essential elements in the control of gene expression
Abstract
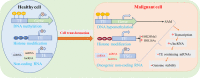
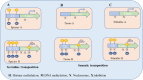
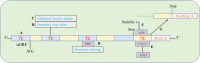
Interspersed repetitions called transposable elements (TEs), commonly referred to as mobile elements, make up a significant portion of the genomes of higher animals. TEs contribute in controlling the expression of genes locally and even far away at the transcriptional and post-transcriptional levels, which is one of their significant functional effects on gene function and genome evolution. There are different mechanisms through which TEs control the expression of genes. First, TEs offer cis-regulatory regions in the genome with their inherent regulatory features for their own expression, making them potential factors for controlling the expression of the host genes. Promoter and enhancer elements contain cis-regulatory sites generated from TE, which function as binding sites for a variety of trans-acting factors. Second, a significant portion of miRNAs and long non-coding RNAs (lncRNAs) have been shown to have TEs that encode for regulatory RNAs, revealing the TE origin of these RNAs. Furthermore, it was shown that TE sequences are essential for these RNAs' regulatory actions, which include binding to the target mRNA. By being a member of cis-regulatory and regulatory RNA sequences, TEs therefore play essential regulatory roles. Additionally, it has been suggested that TE-derived regulatory RNAs and cis-regulatory regions both contribute to the evolutionary novelty of gene regulation. Additionally, these regulatory systems arising from TE frequently have tissue-specific functions. The objective of this review is to discuss TE-mediated gene regulation, with a particular emphasis on the processes, contributions of various TE types, differential roles of various tissue types, based mostly on recent studies on humans.
Keywords: Gene expression; Mobile elements; Transposable elements.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Role of Transposable Elements in Gene Regulation in the Human Genome.Life (Basel). 2021 Feb 4;11(2):118. doi: 10.3390/life11020118. Life (Basel). 2021. PMID: 33557056 Free PMC article. Review.
-
Roles of transposable elements in the regulation of mammalian transcription.Nat Rev Mol Cell Biol. 2022 Jul;23(7):481-497. doi: 10.1038/s41580-022-00457-y. Epub 2022 Feb 28. Nat Rev Mol Cell Biol. 2022. PMID: 35228718 Free PMC article. Review.
-
Molecular Mimicry of Transposable Elements in Plants.Plant Cell Physiol. 2025 May 17;66(4):490-495. doi: 10.1093/pcp/pcae058. Plant Cell Physiol. 2025. PMID: 38808931 Free PMC article. Review.
-
Nearby transposable elements impact plant stress gene regulatory networks: a meta-analysis in A. thaliana and S. lycopersicum.BMC Genomics. 2022 Jan 4;23(1):18. doi: 10.1186/s12864-021-08215-8. BMC Genomics. 2022. PMID: 34983397 Free PMC article.
-
Statistical learning quantifies transposable element-mediated cis-regulation.Genome Biol. 2023 Nov 10;24(1):258. doi: 10.1186/s13059-023-03085-7. Genome Biol. 2023. PMID: 37950299 Free PMC article.
Cited by
-
Association of a 7.9 kb Endogenous Retrovirus Insertion in Intron 1 of CD36 with Obesity and Fat Measurements in Sheep.Mob DNA. 2025 Mar 14;16(1):12. doi: 10.1186/s13100-025-00349-w. Mob DNA. 2025. PMID: 40087777 Free PMC article.
-
Beyond Transposons: TIGD1 as a Pan-Cancer Biomarker and Immune Modulator.Genes (Basel). 2025 May 30;16(6):674. doi: 10.3390/genes16060674. Genes (Basel). 2025. PMID: 40565566 Free PMC article.
-
Long-read RNA sequencing of transposable elements from single cells using CELLO-seq.Nat Protoc. 2025 Jul 16:10.1038/s41596-025-01203-2. doi: 10.1038/s41596-025-01203-2. Online ahead of print. Nat Protoc. 2025. PMID: 40670609 Free PMC article. Review.
-
Update on the Basic Understanding of Fusarium graminearum Virulence Factors in Common Wheat Research.Plants (Basel). 2024 Apr 22;13(8):1159. doi: 10.3390/plants13081159. Plants (Basel). 2024. PMID: 38674569 Free PMC article. Review.
-
Differential Transcriptomic Profile of Piscirickettsia salmonis LF-89 and EM-90 During an In Vivo Spatial Separation Co-Culture in Atlantic Salmon.Microorganisms. 2024 Dec 2;12(12):2480. doi: 10.3390/microorganisms12122480. Microorganisms. 2024. PMID: 39770683 Free PMC article.
References
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

