RaMALDI: Enabling simultaneous Raman and MALDI imaging of the same tissue section
- PMID: 37597501
- PMCID: PMC10544780
- DOI: 10.1016/j.bios.2023.115597
RaMALDI: Enabling simultaneous Raman and MALDI imaging of the same tissue section
Abstract
Multimodal tissue imaging techniques that integrate two complementary modalities are powerful discovery tools for unraveling biological processes and identifying biomarkers of disease. Combining Raman spectroscopic imaging (RSI) and matrix-assisted laser-desorption/ionization (MALDI) mass spectrometry imaging (MSI) to obtain fused images with the advantages of both modalities has the potential of providing spatially resolved, sensitive, specific biomolecular information, but has so far involved two separate sample preparations, or even consecutive tissue sections for RSI and MALDI MSI, resulting in images with inherent disparities. We have developed RaMALDI, a streamlined, integrated, multimodal imaging workflow of RSI and MALDI MSI, performed on a single tissue section with one sample preparation protocol. We show that RaMALDI imaging of various tissues effectively integrates molecular information acquired from both RSI and MALDI MSI of the same sample, which will drive discoveries in cell biology, biomedicine, and pathology, and advance tissue diagnostics.
Keywords: MALDI; Mass spectrometry imaging; Matrix; Multimodal imaging; Raman spectroscopy.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
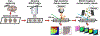




References
-
- Ahlf DR, Masyuko RN, Hummon AB, Bohn PW, 2014. Correlated mass spectrometry imaging and confocal Raman microscopy for studies of three-dimensional cell culture sections. Analyst 139(18), 4578–4585. - PubMed
-
- Ali A, Abouleila Y, Germond A, 2020. An integrated Raman spectroscopy and mass spectrometry platform to study single-cell drug uptake, metabolism, and effects. JoVE (155), e60449. - PubMed
-
- Barry JA, Groseclose MR, Castellino S, 2019. Quantification and assessment of detection capability in imaging mass spectrometry using a revised mimetic tissue model. Bioanalysis 11(11), 1099–1116. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

