APOGEE 2: multi-layer machine-learning model for the interpretable prediction of mitochondrial missense variants
- PMID: 37598215
- PMCID: PMC10439926
- DOI: 10.1038/s41467-023-40797-7
APOGEE 2: multi-layer machine-learning model for the interpretable prediction of mitochondrial missense variants
Abstract
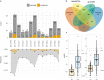
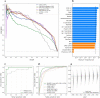
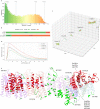
Mitochondrial dysfunction has pleiotropic effects and is frequently caused by mitochondrial DNA mutations. However, factors such as significant variability in clinical manifestations make interpreting the pathogenicity of variants in the mitochondrial genome challenging. Here, we present APOGEE 2, a mitochondrially-centered ensemble method designed to improve the accuracy of pathogenicity predictions for interpreting missense mitochondrial variants. Built on the joint consensus recommendations by the American College of Medical Genetics and Genomics/Association for Molecular Pathology, APOGEE 2 features an improved machine learning method and a curated training set for enhanced performance metrics. It offers region-wise assessments of genome fragility and mechanistic analyses of specific amino acids that cause perceptible long-range effects on protein structure. With clinical and research use in mind, APOGEE 2 scores and pathogenicity probabilities are precompiled and available in MitImpact. APOGEE 2's ability to address challenges in interpreting mitochondrial missense variants makes it an essential tool in the field of mitochondrial genetics.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures
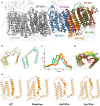

Similar articles
-
Machine learning classifier for identification of damaging missense mutations exclusive to human mitochondrial DNA-encoded polypeptides.BMC Bioinformatics. 2017 Mar 7;18(1):158. doi: 10.1186/s12859-017-1562-7. BMC Bioinformatics. 2017. PMID: 28270093 Free PMC article.
-
MitImpact: an exhaustive collection of pre-computed pathogenicity predictions of human mitochondrial non-synonymous variants.Hum Mutat. 2015 Feb;36(2):E2413-22. doi: 10.1002/humu.22720. Epub 2014 Dec 17. Hum Mutat. 2015. PMID: 25516408
-
Gene-specific machine learning for pathogenicity prediction of rare BRCA1 and BRCA2 missense variants.Sci Rep. 2023 Jun 28;13(1):10478. doi: 10.1038/s41598-023-37698-6. Sci Rep. 2023. PMID: 37380723 Free PMC article.
-
Opening the Black Box: Interpretable Machine Learning for Geneticists.Trends Genet. 2020 Jun;36(6):442-455. doi: 10.1016/j.tig.2020.03.005. Epub 2020 Apr 17. Trends Genet. 2020. PMID: 32396837 Review.
-
Model performance and interpretability of semi-supervised generative adversarial networks to predict oncogenic variants with unlabeled data.BMC Bioinformatics. 2023 Feb 9;24(1):43. doi: 10.1186/s12859-023-05141-2. BMC Bioinformatics. 2023. PMID: 36759776 Free PMC article. Review.
Cited by
-
Mitochondrial DNA variant detection in over 6,500 rare disease families by the systematic analysis of exome and genome sequencing data resolves undiagnosed cases.HGG Adv. 2025 Jul 10;6(3):100441. doi: 10.1016/j.xhgg.2025.100441. Epub 2025 Apr 15. HGG Adv. 2025. PMID: 40241304 Free PMC article.
-
Mitochondrial and Nuclear DNA Variants in Amyotrophic Lateral Sclerosis: Enrichment in the Mitochondrial Control Region and Sirtuin Pathway Genes in Spinal Cord Tissue.Biomolecules. 2024 Mar 28;14(4):411. doi: 10.3390/biom14040411. Biomolecules. 2024. PMID: 38672428 Free PMC article.
-
mtDNA-Server 2: advancing mitochondrial DNA analysis through highly parallelized data processing and interactive analytics.Nucleic Acids Res. 2024 Jul 5;52(W1):W102-W107. doi: 10.1093/nar/gkae296. Nucleic Acids Res. 2024. PMID: 38709886 Free PMC article.
-
Mitochondrial DNA variants revealed by whole exome sequencing: from screening to diagnosis and follow-up.Neurogenetics. 2025 Mar 26;26(1):38. doi: 10.1007/s10048-025-00820-z. Neurogenetics. 2025. PMID: 40138026
-
Our current understanding of the biological impact of endometrial cancer mtDNA genome mutations and their potential use as a biomarker.Front Oncol. 2024 Jun 27;14:1394699. doi: 10.3389/fonc.2024.1394699. eCollection 2024. Front Oncol. 2024. PMID: 38993645 Free PMC article. Review.
References
-
- Muller HJ. The relation of recombination to mutational advance. Mutat. Res. 1964;106:2–9. - PubMed
-
- Allio R, Donega S, Galtier N, Nabholz B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 2017;34:2762–2772. - PubMed
-
- Szczepanowska K, Trifunovic A. Different faces of mitochondrial DNA mutators. Biochim. Biophys. Acta. 2015;1847:1362–1372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

