Inter-comparison of marine microbiome sampling protocols
- PMID: 37598259
- PMCID: PMC10439934
- DOI: 10.1038/s43705-023-00278-w
Inter-comparison of marine microbiome sampling protocols
Abstract
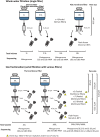
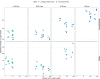
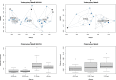
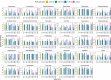
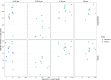
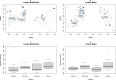
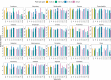
Research on marine microbial communities is growing, but studies are hard to compare because of variation in seawater sampling protocols. To help researchers in the inter-comparison of studies that use different seawater sampling methodologies, as well as to help them design future sampling campaigns, we developed the EuroMarine Open Science Exploration initiative (EMOSE). Within the EMOSE framework, we sampled thousands of liters of seawater from a single station in the NW Mediterranean Sea (Service d'Observation du Laboratoire Arago [SOLA], Banyuls-sur-Mer), during one single day. The resulting dataset includes multiple seawater processing approaches, encompassing different material-type kinds of filters (cartridge membrane and flat membrane), three different size fractionations (>0.22 µm, 0.22-3 µm, 3-20 µm and >20 µm), and a number of different seawater volumes ranging from 1 L up to 1000 L. We show that the volume of seawater that is filtered does not have a significant effect on prokaryotic and protist diversity, independently of the sequencing strategy. However, there was a clear difference in alpha and beta diversity between size fractions and between these and "whole water" (with no pre-fractionation). Overall, we recommend care when merging data from datasets that use filters of different pore size, but we consider that the type of filter and volume should not act as confounding variables for the tested sequencing strategies. To the best of our knowledge, this is the first time a publicly available dataset effectively allows for the clarification of the impact of marine microbiome methodological options across a wide range of protocols, including large-scale variations in sampled volume.
© 2023. ISME Publications B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

