VHH CDR-H3 conformation is determined by VH germline usage
- PMID: 37598276
- PMCID: PMC10439903
- DOI: 10.1038/s42003-023-05241-y
VHH CDR-H3 conformation is determined by VH germline usage
Abstract
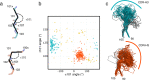
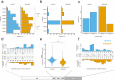
VHHs or nanobodies are single antigen binding domains originating from camelid heavy-chain antibodies. They are used as diagnostic and research tools and in a variety of therapeutic molecules. Analyzing variable domain structures from llama and alpaca we found that VHHs can be classified into two large structural clusters based on their CDR-H3 conformation. Extended CDR-H3 loops protrude into the solvent, whereas kinked CDR-H3 loops fold back onto framework regions. Both major families have distinct properties in terms of their CDR-H3 secondary structure, how their CDR-H3 interacts with the framework region and how they bind to antigens. We show that the CDR-H3 conformation of VHHs correlates with the germline from which the antibodies are derived: IGHV3-3 derived antibodies almost exclusively adopt a kinked CDR-H3 conformation while the CDR-H3 adopts an extended structure in most IGHV3S53 derived antibodies. We do not observe any bias stemming from V(D)J recombination in llama immune repertoires, suggesting that the correlation is the result of selection processes during B-cell development. Our findings demonstrate a previously undescribed impact of germline usage on antigen interaction and contribute to a better understanding on how properties of the antibody framework shape the immune repertoire.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare the following competing interests: All authors are employees of 23andMe Inc.
Figures




References
-
- Austin, R. J. et al. TriTACs, a novel class of T cell-engaging protein constructs designed for the treatment of solid tumors. Mol. Cancer Ther. 109–120 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

