Environmental carcinogens disproportionally mutate genes implicated in neurodevelopmental disorders
- PMID: 37599994
- PMCID: PMC10435087
- DOI: 10.3389/fnins.2023.1106573
Environmental carcinogens disproportionally mutate genes implicated in neurodevelopmental disorders
Abstract
Introduction: De novo mutations contribute to a large proportion of sporadic psychiatric and developmental disorders, yet the potential role of environmental carcinogens as drivers of causal de novo mutations in neurodevelopmental disorders is poorly studied.
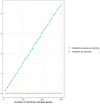
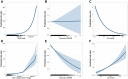
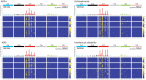
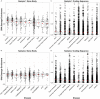
Methods: To explore environmental mutation vulnerability of disease-associated gene sets, we analyzed publicly available whole genome sequencing datasets of mutations in human induced pluripotent stem cell clonal lines exposed to 12 classes of environmental carcinogens, and human lung cancers from individuals living in highly polluted regions. We compared observed rates of exposure-induced mutations in disease-related gene sets with the expected rates of mutations based on control genes randomly sampled from the genome using exact binomial tests. To explore the role of sequence characteristics in mutation vulnerability, we modeled the effects of sequence length, gene expression, and percent GC content on mutation rates of entire genes and gene coding sequences using multivariate Quasi-Poisson regressions.
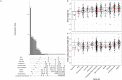
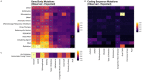
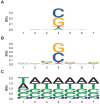
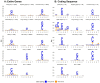
Results: We demonstrate that several mutagens, including radiation and polycyclic aromatic hydrocarbons, disproportionately mutate genes related to neurodevelopmental disorders including autism spectrum disorders, schizophrenia, and attention deficit hyperactivity disorder. Other disease genes including amyotrophic lateral sclerosis, Alzheimer's disease, congenital heart disease, orofacial clefts, and coronary artery disease were generally not mutated more than expected. Longer sequence length was more strongly associated with elevated mutations in entire genes compared with mutations in coding sequences. Increased expression was associated with decreased coding sequence mutation rate, but not with the mutability of entire genes. Increased GC content was associated with increased coding sequence mutation rates but decreased mutation rates in entire genes.
Discussion: Our findings support the possibility that neurodevelopmental disorder genetic etiology is partially driven by a contribution of environment-induced germ line and somatic mutations.
Keywords: autism; carcinogen; de novo mutation; mutagenesis; neurodevelopmental disorders; somatic mutation.
Copyright © 2023 Baker, Zhang, Simon, McLarnan, Chung and Pearson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Association T.A. (2019). Genetics [online]. Washington, DC: The ALS Association.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

