Strategies for Overcoming the Single-Molecule Concentration Barrier
- PMID: 37600457
- PMCID: PMC10436376
- DOI: 10.1021/acsmeasuresciau.3c00002
Strategies for Overcoming the Single-Molecule Concentration Barrier
Abstract
Fluorescence-based single-molecule approaches have helped revolutionize our understanding of chemical and biological mechanisms. Unfortunately, these methods are only suitable at low concentrations of fluorescent molecules so that single fluorescent species of interest can be successfully resolved beyond background signal. The application of these techniques has therefore been limited to high-affinity interactions despite most biological and chemical processes occurring at much higher reactant concentrations. Fortunately, recent methodological advances have demonstrated that this concentration barrier can indeed be broken, with techniques reaching concentrations as high as 1 mM. The goal of this Review is to discuss the challenges in performing single-molecule fluorescence techniques at high-concentration, offer applications in both biology and chemistry, and highlight the major milestones that shatter the concentration barrier. We also hope to inspire the widespread use of these techniques so we can begin exploring the new physical phenomena lying beyond this barrier.
© 2023 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

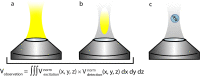
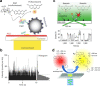
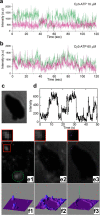


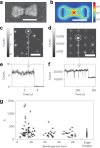
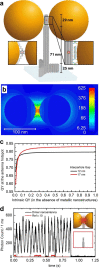
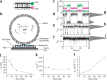
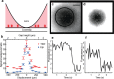


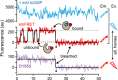
References
-
- Moerner W. E.; Fromm D. P. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev. Sci. Instrum. 2003, 74 (8), 3597–3619. 10.1063/1.1589587. - DOI
-
- Brooks Shera E.; Seitzinger N. K.; Davis L. M.; Keller R. A.; Soper S. A. DETECTION OF SINGLE FLUORESCENT MOLECULES. Chem. Phys. Lett. 1990, 174 (6), 553–557. 10.1016/0009-2614(90)85485-U. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
