POU6F2 mutation in humans with pubertal failure alters GnRH transcript expression
- PMID: 37600690
- PMCID: PMC10436210
- DOI: 10.3389/fendo.2023.1203542
POU6F2 mutation in humans with pubertal failure alters GnRH transcript expression
Abstract
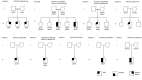
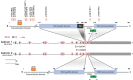
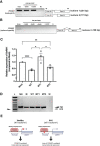
Idiopathic hypogonadotropic hypogonadism (IHH) is characterized by the absence of pubertal development and subsequent impaired fertility often due to gonadotropin-releasing hormone (GnRH) deficits. Exome sequencing of two independent cohorts of IHH patients identified 12 rare missense variants in POU6F2 in 15 patients. POU6F2 encodes two distinct isoforms. In the adult mouse, expression of both isoform1 and isoform2 was detected in the brain, pituitary, and gonads. However, only isoform1 was detected in mouse primary GnRH cells and three immortalized GnRH cell lines, two mouse and one human. To date, the function of isoform2 has been verified as a transcription factor, while the function of isoform1 has been unknown. In the present report, bioinformatics and cell assays on a human-derived GnRH cell line reveal a novel function for isoform1, demonstrating it can act as a transcriptional regulator, decreasing GNRH1 expression. In addition, the impact of the two most prevalent POU6F2 variants, identified in five IHH patients, that were located at/or close to the DNA-binding domain was examined. Notably, one of these mutations prevented the repression of GnRH transcripts by isoform1. Normally, GnRH transcription increases as GnRH cells mature as they near migrate into the brain. Augmentation earlier during development can disrupt normal GnRH cell migration, consistent with some POU6F2 variants contributing to the IHH pathogenesis.
Keywords: GnRH; POU6f2 isoform1; idiopathic hypogonadotropic hypogonadism; puberty; transcription.
Copyright © 2023 Cho, Gurbuz, Stamou, Kotan, Farmer, Can, Tompkins, Mammadova, Altincik, Gokce, Catli, Bugrul, Bartlett, Turan, Balasubramanian, Yuksel, Seminara, Wray and Topaloglu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

