Transcriptional and proteomic analysis of the innate immune response to microbial stimuli in a model invertebrate chordate
- PMID: 37600818
- PMCID: PMC10433773
- DOI: 10.3389/fimmu.2023.1217077
Transcriptional and proteomic analysis of the innate immune response to microbial stimuli in a model invertebrate chordate
Abstract
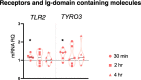
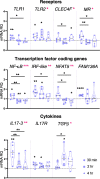
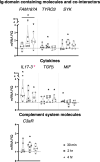
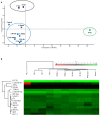
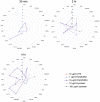
Inflammatory response triggered by innate immunity can act to protect against microorganisms that behave as pathogens, with the aim to restore the homeostatic state between host and beneficial microbes. As a filter-feeder organism, the ascidian Ciona robusta is continuously exposed to external microbes that may be harmful under some conditions. In this work, we used transcriptional and proteomic approaches to investigate the inflammatory response induced by stimuli of bacterial (lipopolysaccharide -LPS- and diacylated lipopeptide - Pam2CSK4) and fungal (zymosan) origin, in Ciona juveniles at stage 4 of metamorphosis. We focused on receptors, co-interactors, transcription factors and cytokines belonging to the TLR and Dectin-1 pathways and on immune factors identified by homology approach (i.e. immunoglobulin (Ig) or C-type lectin domain containing molecules). While LPS did not induce a significant response in juvenile ascidians, Pam2CSK4 and zymosan exposure triggered the activation of specific inflammatory mechanisms. In particular, Pam2CSK4-induced inflammation was characterized by modulation of TLR and Dectin-1 pathway molecules, including receptors, transcription factors, and cytokines, while immune response to zymosan primarily involved C-type lectin receptors, co-interactors, Ig-containing molecules, and cytokines. A targeted proteomic analysis enabled to confirm transcriptional data, also highlighting a temporal delay between transcriptional induction and protein level changes. Finally, a protein-protein interaction network of Ciona immune molecules was rendered to provide a wide visualization and analysis platform of innate immunity. The in vivo inflammatory model described here reveals interconnections of innate immune pathways in specific responses to selected microbial stimuli. It also represents the starting point for studying ontogeny and regulation of inflammatory disorders in different physiological conditions.
Keywords: Ciona robusta; inflammatory response; innate immunity; marine invertebrates; microbial stimuli; protein-protein interactome; proteomic analysis; transcriptional analysis.
Copyright © 2023 Liberti, Pollastro, Pinto, Illiano, Marino, Amoresano, Spagnuolo and Sordino.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

