LIDER: cell embedding based deep neural network classifier for supervised cell type identification
- PMID: 37601262
- PMCID: PMC10439717
- DOI: 10.7717/peerj.15862
LIDER: cell embedding based deep neural network classifier for supervised cell type identification
Abstract
Background: Automatic cell type identification has been an urgent task for the rapid development of single-cell RNA-seq techniques. Generally, the current approach for cell type identification is to generate cell clusters by unsupervised clustering and later assign labels to each cell cluster with manual annotation.
Methods: Here, we introduce LIDER (celL embeddIng based Deep nEural netwoRk classifier), a deep supervised learning method that combines cell embedding and deep neural network classifier for automatic cell type identification. Based on a stacked denoising autoencoder with a tailored and reconstructed loss function, LIDER identifies cell embedding and predicts cell types with a deep neural network classifier. LIDER was developed upon a stacked denoising autoencoder to learn encoder-decoder structures for identifying cell embedding.
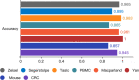
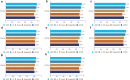
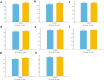
Results: LIDER accurately identifies cell types by using stacked denoising autoencoder. Benchmarking against state-of-the-art methods across eight types of single-cell data, LIDER achieves comparable or even superior enhancement performance. Moreover, LIDER suggests comparable robust to batch effects. Our results show a potential in deep supervised learning for automatic cell type identification of single-cell RNA-seq data. The LIDER codes are available at https://github.com/ShiMGLab/LIDER.
Keywords: Cell embedding; Cell type identification; Deep neural network classifier; Stacked denoising autoencoders.
©2023 Tang et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nature Biotechnology. 2015;33(2):155–160. doi: 10.1038/nbt.3102. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

