The correlation of the intestinal with pharyngeal microbiota in early neonates
- PMID: 37601350
- PMCID: PMC10434775
- DOI: 10.3389/fmicb.2023.1225352
The correlation of the intestinal with pharyngeal microbiota in early neonates
Abstract
Introduction: The gut-lung axis has long been recognized as an important mechanism affecting intestinal and lung immunity. Still, few studies have examined the correlation between the intestinal and pharyngeal microbiota in early neonates, especially when feeding patterns are one of the main drivers of microbiota development.
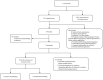
Methods: To explore the composition and function of intestinal and pharyngeal microbiota and to analyze the effect of limited formula feeding on the initial microbiota colonization in early full-term neonates, we characterized the stool and oropharyngeal microbiota of 20 healthy full-term newborns sampled on days 0 and 5-7 after birth using 16S rRNA gene sequencing. Based on the sequencing results, a comparison was made of the compositions and functions of the intestinal and oropharyngeal microbiota for analysis.
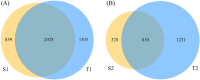
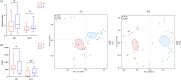
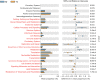
Results and discussion: At the phylum level, Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes were the most abundant in both niches. At the genus level, the species of pioneer bacteria were rich in the intestine and oropharynx but low in abundance on day 0. On days 5-7, Bifidobacterium (25.40%) and Escherichia-Shigella (22.16%) were dominant in the intestine, while Streptococcus (38.40%) and Staphylococcus (23.13%) were dominant in the oropharynx. There were eight core bacteria genera in the intestine and oropharynx on days 5-7, which were Bifidobacterium, Escherichia-Shigella, Staphylococcus, Streptococcus, Bacteroides, Parabacteroides, Rothia, and Acinetobacter. As indicated by PICRUSt analysis, on days 5-7, the intestinal microbiota was more predictive than the oropharyngeal microbiota in transcription, metabolism, cell motility, cellular processes and signaling, and organismal system function in the KEGG pathway. Compared to exclusive breastfeeding, limited formula feeding (40-60%) had no significant effect on the neonatal intestinal and oropharyngeal microbiota composition during the initial colonization period. Our results suggest that the initial colonization of microbiota is closely related to the ecological niche environment in the intestine and oropharynx, with their core microbiota being closely correlated. We found that early limited formula feeding could not significantly affect the initial colonization of microbiota in the intestine and oropharynx.
Keywords: 16S rRNA sequencing; gut-lung axis; intestinal microbiota; neonate; oropharyngeal microbiota.
Copyright © 2023 Wang, Shao, Zhu, Li, You and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
[Intestinal and pharyngeal microbiota in early neonates: an analysis based on high-throughput sequencing].Zhongguo Dang Dai Er Ke Za Zhi. 2023 May 15;25(5):508-515. doi: 10.7499/j.issn.1008-8830.2301015. Zhongguo Dang Dai Er Ke Za Zhi. 2023. PMID: 37272178 Free PMC article. Chinese.
-
Clinical Study of Correlation for the Intestinal and Pharyngeal Microbiota in the Premature Neonates.Front Pediatr. 2021 Feb 16;9:632573. doi: 10.3389/fped.2021.632573. eCollection 2021. Front Pediatr. 2021. PMID: 33665178 Free PMC article.
-
Distribution characteristics of oral microbiota and its relationship with intestinal microbiota in patients with type 2 diabetes mellitus.Front Endocrinol (Lausanne). 2023 Mar 16;14:1119201. doi: 10.3389/fendo.2023.1119201. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37025407 Free PMC article.
-
A pilot study on the characterization and correlation of oropharyngeal and intestinal microbiota in children with type 1 diabetes mellitus.Front Pediatr. 2024 Jun 13;12:1382466. doi: 10.3389/fped.2024.1382466. eCollection 2024. Front Pediatr. 2024. PMID: 38938502 Free PMC article.
-
Gut Microbiota and Its Repercussion in Parkinson's Disease: A Systematic Review in Occidental Patients.Neurol Int. 2023 Jun 13;15(2):750-763. doi: 10.3390/neurolint15020047. Neurol Int. 2023. PMID: 37368331 Free PMC article. Review.
Cited by
-
Comparison of intestinal and pharyngeal microbiota in preterm infants on the first day of life and the characteristics of pharyngeal microbiota in infants delivered by cesarean section or vaginally.Front Pediatr. 2024 Oct 8;12:1411887. doi: 10.3389/fped.2024.1411887. eCollection 2024. Front Pediatr. 2024. PMID: 39439450 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

