Trade-off between local replication and long-distance dissemination during experimental evolution of a satellite RNA
- PMID: 37601360
- PMCID: PMC10436602
- DOI: 10.3389/fmicb.2023.1139447
Trade-off between local replication and long-distance dissemination during experimental evolution of a satellite RNA
Abstract
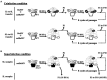
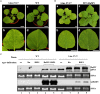
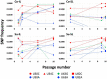
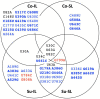
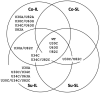
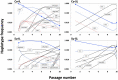
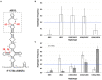
Satellite RNAs (satRNAs) are molecular parasites that depend on their non-homologous helper viruses (HVs) for essential biological functions. While there are multiple molecular and phylogenetic studies on satRNAs, there is no experimental evolution study on how satRNAs may evolve in common infection conditions. In this study, we serially passaged the Bamboo mosaic virus (BaMV) associated-satRNA (satBaMV) under conditions in which satBaMV either coinfects an uninfected host plant, Nicotiana benthamiana, with BaMV or superinfects a transgenic N. benthamiana expressing the full-length BaMV genome. Single-nucleotide polymorphisms (SNPs) of satBaMV populations were analyzed by deep sequencing. Forty-eight SNPs were identified across four different experimental treatments. Most SNPs are treatment-specific, and some are also ephemeral. However, mutations at positions 30, 34, 63, and 82, all located at the 5' untranslated region (UTR), are universal in all treatments. These universal SNPs are configured into several haplotypes and follow different population dynamics. We constructed isogenic satBaMV strains only differing at positions 30 and 82 and conducted competition experiments in protoplasts and host plants. We found that the haplotype that reached high frequency in protoplasts and inoculation leaves also exhibited poor dissemination to systemic leaves and vice versa, thus suggesting an apparent trade-off between local replication and long-distance dissemination. We posit that the trade-off is likely caused by antagonistic pleiotropy at the 5' UTR. Our findings revealed a hitherto under-explored connection between satRNA genome replication and movement within a host plant. The significance of such a connection during satRNA evolution warrants a more thorough investigation.
Keywords: antagonistic pleiotropy; bamboo mosaic virus (BaMV); experimental evolution; satBaMV; satellite RNA (satRNA); serial passage.
Copyright © 2023 Lee, Liou, Hsu, Wang and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Bawden F. C., Pirie N. W. (1942). A preliminary description of preparations of some of the viruses causing tobacco necrosis. Brit. J. Exp. Path. 23, 314–327.
LinkOut - more resources
Full Text Sources

