A critical analysis of the current state of virus taxonomy
- PMID: 37601376
- PMCID: PMC10435761
- DOI: 10.3389/fmicb.2023.1240993
A critical analysis of the current state of virus taxonomy
Abstract
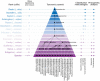
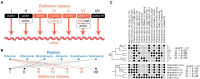
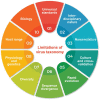
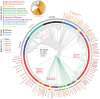
Taxonomical classification has preceded evolutionary understanding. For that reason, taxonomy has become a battleground fueled by knowledge gaps, technical limitations, and a priorism. Here we assess the current state of the challenging field, focusing on fallacies that are common in viral classification. We emphasize that viruses are crucial contributors to the genomic and functional makeup of holobionts, organismal communities that behave as units of biological organization. Consequently, viruses cannot be considered taxonomic units because they challenge crucial concepts of organismality and individuality. Instead, they should be considered processes that integrate virions and their hosts into life cycles. Viruses harbor phylogenetic signatures of genetic transfer that compromise monophyly and the validity of deep taxonomic ranks. A focus on building phylogenetic networks using alignment-free methodologies and molecular structure can help mitigate the impasse, at least in part. Finally, structural phylogenomic analysis challenges the polyphyletic scenario of multiple viral origins adopted by virus taxonomy, defeating a polyphyletic origin and supporting instead an ancient cellular origin of viruses. We therefore, prompt abandoning deep ranks and urgently reevaluating the validity of taxonomic units and principles of virus classification.
Keywords: ICTV; classification; evolution; holobiont; horizontal genetic transfer; reticulation; virus origin.
Copyright © 2023 Caetano-Anollés, Claverie and Nasir.
Conflict of interest statement
AN is a shareholder and employee at Moderna, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures