Touching the (almost) untouchable: a minimally invasive workflow for microbiological and biomolecular analyses of cultural heritage objects
- PMID: 37601377
- PMCID: PMC10435870
- DOI: 10.3389/fmicb.2023.1197837
Touching the (almost) untouchable: a minimally invasive workflow for microbiological and biomolecular analyses of cultural heritage objects
Abstract
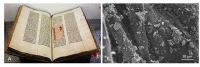
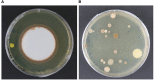
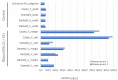
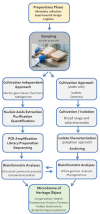
Microbiological and biomolecular approaches to cultural heritage research have expanded the established research horizon from the prevalent focus on the cultural objects' conservation and human health protection to the relatively recent applications to provenance inquiry and assessment of environmental impacts in a global context of a changing climate. Standard microbiology and molecular biology methods developed for other materials, specimens, and contexts could, in principle, be applied to cultural heritage research. However, given certain characteristics common to several heritage objects-such as uniqueness, fragility, high value, and restricted access, tailored approaches are required. In addition, samples of heritage objects may yield low microbial biomass, rendering them highly susceptible to cross-contamination. Therefore, dedicated methodology addressing these limitations and operational hurdles is needed. Here, we review the main experimental challenges and propose a standardized workflow to study the microbiome of cultural heritage objects, illustrated by the exploration of bacterial taxa. The methodology was developed targeting the challenging side of the spectrum of cultural heritage objects, such as the delicate written record, while retaining flexibility to adapt and/or upscale it to heritage artifacts of a more robust constitution or larger dimensions. We hope this tailored review and workflow will facilitate the interdisciplinary inquiry and interactions among the cultural heritage research community.
Keywords: 16S rRNA gene; archaeological artifacts; biocodicology; cultural heritage; medieval manuscripts; microbial signatures; microbiome; minimally invasive.
Copyright © 2023 Flocco, Methner, Burkart, Geppert and Overmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abel M.-L. (2011). Surface and interface analysis in cultural heritage. Surf. Interface Anal. 43, 1099–1099. 10.1002/sia.3805 - DOI
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed July 24, 2023).
LinkOut - more resources
Full Text Sources

