Molecular residual disease detection in resected, muscle-invasive urothelial cancer with a tissue-based comprehensive genomic profiling-informed personalized monitoring assay
- PMID: 37601688
- PMCID: PMC10433150
- DOI: 10.3389/fonc.2023.1221718
Molecular residual disease detection in resected, muscle-invasive urothelial cancer with a tissue-based comprehensive genomic profiling-informed personalized monitoring assay
Abstract
Introduction: Circulating tumor DNA (ctDNA) detection postoperatively may identify patients with urothelial cancer at a high risk of relapse. Pragmatic tools building off clinical tumor next-generation sequencing (NGS) platforms could have the potential to increase assay accessibility.
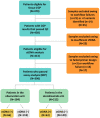
Methods: We evaluated the widely available Foundation Medicine comprehensive genomic profiling (CGP) platform as a source of variants for tracking of ctDNA when analyzing residual samples from IMvigor010 (ClinicalTrials.gov identifier NCT02450331), a randomized adjuvant study comparing atezolizumab with observation after bladder cancer surgery. Current methods often involve germline sampling, which is not always feasible or practical. Rather than performing white blood cell sequencing to filter germline and clonal hematopoiesis (CH) variants, we applied a bioinformatic approach to select tumor (non-germline/CH) variants for molecular residual disease detection. Tissue-informed personalized multiplex polymerase chain reaction-NGS assay was used to detect ctDNA postsurgically (Natera).
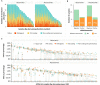
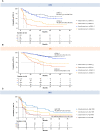
Results: Across 396 analyzed patients, prevalence of potentially actionable alterations was comparable with the expected prevalence in advanced disease (13% FGFR2/3, 20% PIK3CA, 13% ERBB2, and 37% with elevated tumor mutational burden ≥10 mutations/megabase). In the observation arm, 66 of the 184 (36%) ctDNA-positive patients had shorter disease-free survival [DFS; hazard ratio (HR) = 5.77; 95% confidence interval (CI), 3.84-8.67; P < 0.0001] and overall survival (OS; HR = 5.81; 95% CI, 3.41-9.91; P < 0.0001) compared with ctDNA-negative patients. ctDNA-positive patients had improved DFS and OS with atezolizumab compared with those in observation (DFS HR = 0.56; 95% CI, 0.38-0.83; P = 0.003; OS HR = 0.66; 95% CI, 0.42-1.05). Clinical sensitivity and specificity for detection of postsurgical recurrence were 58% (60/103) and 93% (75/81), respectively.
Conclusion: We present a personalized ctDNA monitoring assay utilizing tissue-based FoundationOne® CDx CGP, which is a pragmatic and potentially clinically scalable method that can detect low levels of residual ctDNA in patients with resected, muscle-invasive bladder cancer without germline sampling.
Keywords: CtDNA; MRD; bladder cancer; comprehensive genomic profiling; immunotherapy; monitoring assay; next-generation sequencing.
Copyright © 2023 Powles, Young, Nimeiri, Madison, Fine, Zollinger, Huang, Xu, Gjoerup, Aushev, Wu, Aleshin, Carter, Davarpanah, Degaonkar, Gupta, Mariathasan, Schleifman, Assaf, Oxnard and Hegde.
Conflict of interest statement
TP received honoraria from advisory/consultancy roles with AstraZeneca, BMS, Exelixis, Incyte, Ipsen, Merck, MSD, Novartis, Pfizer, Seattle Genetics, Merck Serono EMD Serono, Astellas, Johnson & Johnson, Eisai, Mashup Ltd, and Roche; institutional research funding support from AstraZeneca, Roche, BMS, Exelixis, Ipsen, Merck, MSD, Novartis, Pfizer, Seattle Genetics, Merck Serono EMD Serono, Astellas, Eisai, and Johnson & Johnson; and travel, accommodation, and expenses support from Roche, Pfizer, MSD, AstraZeneca, and Ipsen. AY, HN, RM, AF, DZ, YH, CX, OG, GO, and PH are employees of Foundation Medicine, a wholly owned subsidiary of Roche, and have equity interest in Roche. VA, HW, and AA are employees of Natera, Inc., and reports stock ownership in Natera. CC, ND, VD, PG, SM, ES, and ZA are employees of Genentech and have equity interest in Roche.
Figures



References
-
- Christensen E, Birkenkamp-Demtröder K, Sethi H, Shchegrova S, Salari R, Nordentoft I, et al. . Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J Clin Oncol (2019) 37(18):1547–57. doi: 10.1200/JCO.18.02052 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

