Identification and interpretation of TET2 noncanonical splicing site intronic variants in myeloid neoplasm patients
- PMID: 37601840
- PMCID: PMC10435687
- DOI: 10.1002/jha2.744
Identification and interpretation of TET2 noncanonical splicing site intronic variants in myeloid neoplasm patients
Abstract
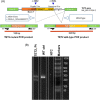
Background: DNA hypermethylation and instability due to inactivation mutations in Ten-eleven translocation 2 (TET2) is a key biomarker of hematological malignancies. This study aims at characterizing two intronic noncanonical splice-site variants, c.3954+5_3954+8delGTTT and c.3954+5G>A. Methods: We used in silico prediction tools, reverse transcription (RT)-PCR, and Sanger sequencing on blood/bone marrow-derived RNA specimens to determine the aberrant splicing. Results: In silico prediction of both variants exhibited reduced splicing strength at the TET2 intron 7 splicing donor site. RT-PCR and Sanger sequencing identified a 62-bp deletion at the exon 7, producing a frameshift mutation, p.Cys1298*. Conclusion: This study provides functional evidence for two intronic TET2 variants that cause alternative splicing and frameshift mutation.
Keywords: TET2; in silico prediction; myeloid neoplasm; next‐generation sequencing (NGS); noncanonical splicing site; ten‐eleven translocation 2; tumor suppressor.
© 2023 The Authors. eJHaem published by British Society for Haematology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Figures



Similar articles
-
Identification of Novel MET Exon 14 Skipping Variants in Non-Small Cell Lung Cancer Patients: A Prototype Workflow Involving in Silico Prediction and RT-PCR.Cancers (Basel). 2022 Oct 1;14(19):4814. doi: 10.3390/cancers14194814. Cancers (Basel). 2022. PMID: 36230737 Free PMC article.
-
Identification of novel point mutations in splicing sites integrating whole-exome and RNA-seq data in myeloproliferative diseases.Mol Genet Genomic Med. 2013 Nov;1(4):246-59. doi: 10.1002/mgg3.23. Epub 2013 Jul 7. Mol Genet Genomic Med. 2013. PMID: 24498620 Free PMC article.
-
Insights of Noncanonical Splice-site Variants on RNA Splicing in Patients With Congenital Hypothyroidism.J Clin Endocrinol Metab. 2022 Feb 17;107(3):e1263-e1276. doi: 10.1210/clinem/dgab737. J Clin Endocrinol Metab. 2022. PMID: 34632506
-
Understanding human DNA variants affecting pre-mRNA splicing in the NGS era.Adv Genet. 2019;103:39-90. doi: 10.1016/bs.adgen.2018.09.002. Epub 2019 Jan 17. Adv Genet. 2019. PMID: 30904096 Review.
-
Rules and tools to predict the splicing effects of exonic and intronic mutations.Wiley Interdiscip Rev RNA. 2018 Jan;9(1). doi: 10.1002/wrna.1451. Epub 2017 Sep 26. Wiley Interdiscip Rev RNA. 2018. PMID: 28949076 Review.
References
-
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–301. - PubMed
LinkOut - more resources
Full Text Sources
