Joint analysis of GWAS and multi-omics QTL summary statistics reveals a large fraction of GWAS signals shared with molecular phenotypes
- PMID: 37601976
- PMCID: PMC10435383
- DOI: 10.1016/j.xgen.2023.100344
Joint analysis of GWAS and multi-omics QTL summary statistics reveals a large fraction of GWAS signals shared with molecular phenotypes
Abstract
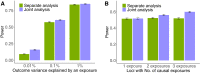
Molecular quantitative trait loci (xQTLs) are often harnessed to prioritize genes or functional elements underpinning variant-trait associations identified from genome-wide association studies (GWASs). Here, we introduce OPERA, a method that jointly analyzes GWAS and multi-omics xQTL summary statistics to enhance the identification of molecular phenotypes associated with complex traits through shared causal variants. Applying OPERA to summary-level GWAS data for 50 complex traits (n = 20,833-766,345) and xQTL data from seven omics layers (n = 100-31,684) reveals that 50% of the GWAS signals are shared with at least one molecular phenotype. GWAS signals shared with multiple molecular phenotypes, such as those at the MSMB locus for prostate cancer, are particularly informative for understanding the genetic regulatory mechanisms underlying complex traits. Future studies with more molecular phenotypes, measured considering spatiotemporal effects in larger samples, are required to obtain a more saturated map linking molecular intermediates to GWAS signals.
Keywords: Bayesian analysis; complex trait; gene discovery; genetic regulatory mechanisms; genome-wide association study; joint analysis; molecular phenotype; molecular quantitative trait locus; multi-omics; summary statistics.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J.T., Loh P.R., Schoech A., Reshef Y., Liu X., O'Connor L., et al. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. - DOI - PMC - PubMed
-
- Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., Schizophrenia Working Group of the Psychiatric Genomics Consortium. McCarroll S., et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

