Transcriptomic forecasting with neural ordinary differential equations
- PMID: 37602211
- PMCID: PMC10435954
- DOI: 10.1016/j.patter.2023.100793
Transcriptomic forecasting with neural ordinary differential equations
Abstract
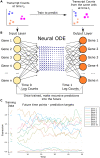
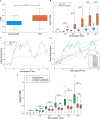
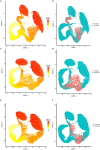
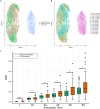
Single-cell transcriptomics technologies can uncover changes in the molecular states that underlie cellular phenotypes. However, understanding the dynamic cellular processes requires extending from inferring trajectories from snapshots of cellular states to estimating temporal changes in cellular gene expression. To address this challenge, we have developed a neural ordinary differential-equation-based method, RNAForecaster, for predicting gene expression states in single cells for multiple future time steps in an embedding-independent manner. We demonstrate that RNAForecaster can accurately predict future expression states in simulated single-cell transcriptomic data with cellular tracking over time. We then show that by using metabolic labeling single-cell RNA sequencing (scRNA-seq) data from constitutively dividing cells, RNAForecaster accurately recapitulates many of the expected changes in gene expression during progression through the cell cycle over a 3-day period. Thus, RNAForecaster enables short-term estimation of future expression states in biological systems from high-throughput datasets with temporal information.
Keywords: artificial intelligence; cellular phenotypes; machine learning; neural ODE; predictive biology; single-cell RNA-seq; temporalomics.
© 2023 The Author(s).
Conflict of interest statement
The corresponding author is on the Scientific Advisory Board of Resistance Bio/Viosera Therapeutics and is a consultant for Mestag Therapeutics and Merck.
Figures

References
-
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. - DOI - PMC - PubMed
-
- Schiebinger G., Shu J., Tabaka M., Cleary B., Subramanian V., Solomon A., Gould J., Liu S., Lin S., Berube P., et al. Optimal-Transport Analysis of Single-Cell Gene Expression Identifies Developmental Trajectories in Reprogramming. Cell. 2019;176:928–943.e22. doi: 10.1016/j.cell.2019.01.006. - DOI - PMC - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

