Interactive analysis of single-cell data using flexible workflows with SCTK2
- PMID: 37602214
- PMCID: PMC10436054
- DOI: 10.1016/j.patter.2023.100814
Interactive analysis of single-cell data using flexible workflows with SCTK2
Abstract
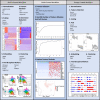
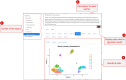
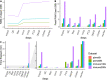
Analysis of single-cell RNA sequencing (scRNA-seq) data can reveal novel insights into the heterogeneity of complex biological systems. Many tools and workflows have been developed to perform different types of analyses. However, these tools are spread across different packages or programming environments, rely on different underlying data structures, and can only be utilized by people with knowledge of programming languages. In the Single-Cell Toolkit 2 (SCTK2), we have integrated a variety of popular tools and workflows to perform various aspects of scRNA-seq analysis. All tools and workflows can be run in the R console or using an intuitive graphical user interface built with R/Shiny. HTML reports generated with Rmarkdown can be used to document and recapitulate individual steps or entire analysis workflows. We show that the toolkit offers more features when compared with existing tools and allows for a seamless analysis of scRNA-seq data for non-computational users.
Keywords: analysis; bioinformatics; genomic; graphical user interface; interactive; interoperability; single cell; software; toolkit; transcriptomic.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Li B., Gould J., Yang Y., Sarkizova S., Tabaka M., Ashenberg O., Rosen Y., Slyper M., Kowalczyk M.S., Villani A.-C., et al. Cumulus provides cloud-based data analysis for large-scale single-cell and single-nucleus RNA-seq. Nat. Methods. 2020;17:793–798. doi: 10.1038/s41592-020-0905-x. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

