Inferring CTCF-binding patterns and anchored loops across human tissues and cell types
- PMID: 37602215
- PMCID: PMC10436006
- DOI: 10.1016/j.patter.2023.100798
Inferring CTCF-binding patterns and anchored loops across human tissues and cell types
Abstract
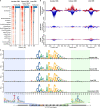
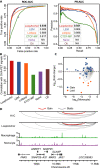
CCCTC-binding factor (CTCF) is a transcription regulator with a complex role in gene regulation. The recognition and effects of CTCF on DNA sequences, chromosome barriers, and enhancer blocking are not well understood. Existing computational tools struggle to assess the regulatory potential of CTCF-binding sites and their impact on chromatin loop formation. Here we have developed a deep-learning model, DeepAnchor, to accurately characterize CTCF binding using high-resolution genomic/epigenomic features. This has revealed distinct chromatin and sequence patterns for CTCF-mediated insulation and looping. An optimized implementation of a previous loop model based on DeepAnchor score excels in predicting CTCF-anchored loops. We have established a compendium of CTCF-anchored loops across 52 human tissue/cell types, and this suggests that genomic disruption of these loops could be a general mechanism of disease pathogenesis. These computational models and resources can help investigate how CTCF-mediated cis-regulatory elements shape context-specific gene regulation in cell development and disease progression.
Keywords: 3D genome; CTCF; CTCF-mediated loop; cis-regulatory element; deep neural networks.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Deep Learning of Sequence Patterns for CCCTC-Binding Factor-Mediated Chromatin Loop Formation.J Comput Biol. 2021 Feb;28(2):133-145. doi: 10.1089/cmb.2020.0225. Epub 2020 Nov 25. J Comput Biol. 2021. PMID: 33232622
-
CLNN-loop: a deep learning model to predict CTCF-mediated chromatin loops in the different cell lines and CTCF-binding sites (CBS) pair types.Bioinformatics. 2022 Sep 30;38(19):4497-4504. doi: 10.1093/bioinformatics/btac575. Bioinformatics. 2022. PMID: 35997565
-
A sequence-based deep learning approach to predict CTCF-mediated chromatin loop.Brief Bioinform. 2021 Sep 2;22(5):bbab031. doi: 10.1093/bib/bbab031. Brief Bioinform. 2021. PMID: 33634313
-
On the choreography of genome folding: A grand pas de deux of cohesin and CTCF.Curr Opin Cell Biol. 2021 Jun;70:84-90. doi: 10.1016/j.ceb.2020.12.001. Epub 2021 Feb 2. Curr Opin Cell Biol. 2021. PMID: 33545664 Review.
-
Many facades of CTCF unified by its coding for three-dimensional genome architecture.J Genet Genomics. 2020 Aug;47(8):407-424. doi: 10.1016/j.jgg.2020.06.008. Epub 2020 Sep 1. J Genet Genomics. 2020. PMID: 33187878 Review.
Cited by
-
CTCF-anchored chromatin loop dynamics during human meiosis.BMC Biol. 2025 Mar 20;23(1):83. doi: 10.1186/s12915-025-02181-3. BMC Biol. 2025. PMID: 40114154 Free PMC article.
References
LinkOut - more resources
Full Text Sources

