Genetic Architecture of Flowering Time Differs Between Populations With Contrasting Demographic and Selective Histories
- PMID: 37603463
- PMCID: PMC10461413
- DOI: 10.1093/molbev/msad185
Genetic Architecture of Flowering Time Differs Between Populations With Contrasting Demographic and Selective Histories
Abstract
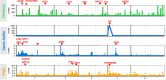
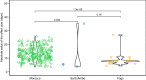
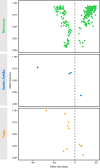
Understanding the evolutionary factors that impact the genetic architecture of traits is a central goal of evolutionary genetics. Here, we investigate how quantitative trait variation accumulated over time in populations that colonized a novel environment. We compare the genetic architecture of flowering time in Arabidopsis populations from the drought-prone Cape Verde Islands and their closest outgroup population from North Africa. We find that trait polygenicity is severely reduced in the island populations compared to the continental North African population. Further, trait architectures and reconstructed allelic histories best fit a model of strong directional selection in the islands in accord with a Fisher-Orr adaptive walk. Consistent with this, we find that large-effect variants that disrupt major flowering time genes (FRI and FLC) arose first, followed by smaller effect variants, including ATX2 L125F, which is associated with a 4-day reduction in flowering time. The most recently arising flowering time-associated loci are not known to be directly involved in flowering time, consistent with an omnigenic signature developing as the population approaches its trait optimum. Surprisingly, we find no effect in the natural population of EDI-Cvi-0 (CRY2 V367M), an allele for which an effect was previously validated by introgression into a Eurasian line. Instead, our results suggest the previously observed effect of the EDI-Cvi-0 allele on flowering time likely depends on genetic background, due to an epistatic interaction. Altogether, our results provide an empirical example of the effects demographic history and selection has on trait architecture.
Keywords: Fisher–Orr geometric model; adaptive walk; complex traits; directional selection; flowering time.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures



References
-
- Ågren J, Oakley CG, Lundemo S, Schemske DW. 2017. Adaptive divergence in flowering time among natural populations of Arabidopsis thaliana: estimates of selection and QTL mapping. Evolution. 71(3):550–564. - PubMed
-
- Ågren J, Schemske DW. 2012. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 194(4):1112–1122. - PubMed
-
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MTR. 1998. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population: AFLP based linkage map of Arabidopsis. Plant J. 14(2):259–271. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

