Prognostic value and potential molecular mechanism of ITGB superfamily members in hepatocellular carcinoma
- PMID: 37603520
- PMCID: PMC10443747
- DOI: 10.1097/MD.0000000000034765
Prognostic value and potential molecular mechanism of ITGB superfamily members in hepatocellular carcinoma
Abstract
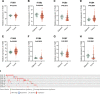
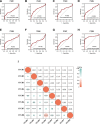
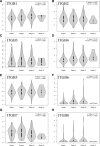
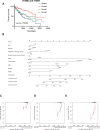
We analyzed the prognostic value and potential molecular mechanisms of the members of integrin β (ITGB)superfamily in hepatocellular carcinoma (HCC) using data from The Cancer Genome Atlas (TCGA), cBioPortal, Gene Expression Profiling Interactive Analysis (GEPIA), Human Protein Atlas (HPA) HPA, Search Tool for the Retrieval of Interacting Genes/Proteins, GeneMANIA, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), TIMER and Gene set enrichment analysis (GSEA) databases. ITGB4/5 mRNA was upregulated in HCC tissues in contrast to the normal liver tissues, whereas ITGB2/3/8 levels were lower in the former. ITGB4 was the most frequently mutated ITGB gene in HCC. Receiver operating characteristic curve (ROC) analysis showed that the expression levels of ITGB2/3/4/5/7/8 had significant diagnostic value in distinguishing HCC tissues from healthy liver tissues, ITGB8 had the highest diagnostic efficacy. The ITGB1/3/6/8 were also upregulated in the HCC tissues in contrast to healthy liver tissues. The expression of ITGB8 was verified by immunohistochemistry (IHC). Furthermore, ITGB6 and ITGB7 expression levels were strongly associated with the overall survival (OS) of HCC patients. The ITGB superfamily members exhibited homology and interactions in protein structure. In addition, ITGB6 together with ITGB7 were negatively related to the infiltration of multiple immune cell populations. GSEA results showed that ITGB6 was enriched in HCC migration and recurrence, whereas ITGB7 was significantly enriched in HIPPO, TOLL and JAK-STAT signaling pathways. In conclusion, ITGB6 and ITGB7 genes are possible to be prognostic biomarkers for HCC.
Copyright © 2023 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.How to cite this article: Xie H, Qin C, Zhou X, Liu J, Yang K, Nong J, Luo J, Peng T. Prognostic value and potential molecular mechanism of ITGB superfamily members in hepatocellular carcinoma. Medicine 2023;102:33(e34765).
Figures






References
-
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:7. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

