LAMP1/2 as potential diagnostic and prognostic marker for brain lower grade glioma: A review
- PMID: 37603525
- PMCID: PMC10443750
- DOI: 10.1097/MD.0000000000034604
LAMP1/2 as potential diagnostic and prognostic marker for brain lower grade glioma: A review
Abstract
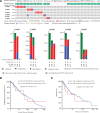
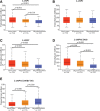
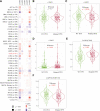
Brain lower grade glioma (LGG) is a common type of glioma. The current treatment methods still have some limitations, and some LGG patients will inevitably continue to deteriorate after treatment. We found the value of lysosomal associated membrane proteins (LAMPs) in the diagnosis and prognosis of LGG, which helps to enhance the clinical understanding of LGG treatment and improved prognosis. We assess the role of LAMPs in LGG, via the publicly available TCGA database. We explored expression levels of LAMPs in LGG using GEPIA2, cBioPortal, and UALCAN databases. The correction of LAMPs expression levels with immune cell infiltration in LGG patient was assessed by TIMER database. The Lysosomal associated membrane protein 1 (LAMP1)/2/4 mRNA levels were significantly higher in LGG patients than in healthy controls. Morover, high mRNA expressions of LAMP1/2/Lysosomal associated membrane protein 3 were associated with poor overall survival. We found that the immune invasion of LGG was almost significantly correlated with the expression of LAMPs. The results suggested that mRNA expressions of LAMP1 and LAMP4 were significantly associated with histological subtypes in LGG patients. lysosomal associated membrane protein 2 and LAMP5 were significantly down-regulated expression in samples of TP53 mutant in LGG compared to TP53 wild type. In addition, Lysosomal associated membrane protein 3 and LAMP4 were significantly overexpressed in samples of TP53 mutant in LGG Enrichment analysis applied to each component indicated that biological function was primarily associated with series of pathways in synapse and immunity.
Copyright © 2023 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures




References
-
- Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

