Widespread infection, diversification and old host associations of Nosema Microsporidia in European freshwater gammarids (Amphipoda)
- PMID: 37603557
- PMCID: PMC10470943
- DOI: 10.1371/journal.ppat.1011560
Widespread infection, diversification and old host associations of Nosema Microsporidia in European freshwater gammarids (Amphipoda)
Abstract
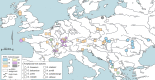
The microsporidian genus Nosema is primarily known to infect insects of economic importance stimulating high research interest, while other hosts remain understudied. Nosema granulosis is one of the formally described Nosema species infecting amphipod crustaceans, being known to infect only two host species. Our first aim was to characterize Nosema spp. infections in different amphipod species from various European localities using the small subunit ribosomal DNA (SSU) marker. Second, we aimed to assess the phylogenetic diversity, host specificity and to explore the evolutionary history that may explain the diversity of gammarid-infecting Nosema lineages by performing a phylogenetic reconstruction based on RNA polymerase II subunit B1 (RPB1) gene sequences. For the host species Gammarus balcanicus, we also analyzed whether parasites were in excess in females to test for sex ratio distortion in relation with Nosema infection. We identified Nosema spp. in 316 individuals from nine amphipod species being widespread in Europe. The RPB1-based phylogenetic reconstruction using newly reported sequences and available data from other invertebrates identified 39 haplogroups being associated with amphipods. These haplogroups clustered into five clades (A-E) that did not form a single amphipod-infecting monophyletic group. Closely related sister clades C and D correspond to Nosema granulosis. Clades A, B and E might represent unknown Nosema species infecting amphipods. Host specificity seemed to be variable with some clades being restricted to single hosts, and some that could be found in several host species. We show that Nosema parasite richness in gammarid hosts is much higher than expected, illustrating the advantage of the use of RPB1 marker over SSU. Finally, we found no hint of sex ratio distortion in Nosema clade A infecting G. balcanicus. This study shows that Nosema spp. are abundant, widespread and diverse in European gammarids. Thus, Nosema is as diverse in aquatic as in terrestrial hosts.
Copyright: © 2023 Bacela-Spychalska et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Franzen C. Microsporidia: A review of 150 years of research. Open Parasitol J. 2008;2: 1–34. doi: 10.2174/1874421400802010001 - DOI
-
- Naegeli K. Über die neue Krankheit der Seidenraupe und verwandte Organismen. Botanische Zeitung. 1857;15: 760–761.
-
- Tokarev YS, Huang W-F, Solter LF, Malysh JM, Becnel JJ, Vossbrinck CR. A formal redefinition of the genera Nosema and Vairimorpha (Microsporidia: Nosematidae) and reassignment of species based on molecular phylogenetics. J Invertebr Pathol. 2020;169: 107279. doi: 10.1016/j.jip.2019.107279 - DOI - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

