Efficient sampling-based Bayesian Active Learning for synaptic characterization
- PMID: 37603559
- PMCID: PMC10470935
- DOI: 10.1371/journal.pcbi.1011342
Efficient sampling-based Bayesian Active Learning for synaptic characterization
Abstract
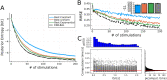
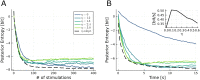
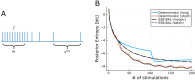
Bayesian Active Learning (BAL) is an efficient framework for learning the parameters of a model, in which input stimuli are selected to maximize the mutual information between the observations and the unknown parameters. However, the applicability of BAL to experiments is limited as it requires performing high-dimensional integrations and optimizations in real time. Current methods are either too time consuming, or only applicable to specific models. Here, we propose an Efficient Sampling-Based Bayesian Active Learning (ESB-BAL) framework, which is efficient enough to be used in real-time biological experiments. We apply our method to the problem of estimating the parameters of a chemical synapse from the postsynaptic responses to evoked presynaptic action potentials. Using synthetic data and synaptic whole-cell patch-clamp recordings, we show that our method can improve the precision of model-based inferences, thereby paving the way towards more systematic and efficient experimental designs in physiology.
Copyright: © 2023 Gontier et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Emery AF, Nenarokomov AV. Optimal experiment design. Measurement Science and Technology. 1998;9(6):864. doi: 10.1088/0957-0233/9/6/003 - DOI
-
- Sebastiani P, Wynn HP. Maximum entropy sampling and optimal Bayesian experimental design. Journal of the Royal Statistical Society: Series B (Statistical Methodology). 2000;62(1):145–157. doi: 10.1111/1467-9868.00225 - DOI

