Base Editor Scanning Reveals Activating Mutations of DNMT3A
- PMID: 37603861
- PMCID: PMC10560492
- DOI: 10.1021/acschembio.3c00257
Base Editor Scanning Reveals Activating Mutations of DNMT3A
Abstract
DNA methyltransferase 3A (DNMT3A) is a de novo cytosine methyltransferase responsible for establishing proper DNA methylation during mammalian development. Loss-of-function (LOF) mutations to DNMT3A, including the hotspot mutation R882H, frequently occur in developmental growth disorders and hematological diseases, including clonal hematopoiesis and acute myeloid leukemia. Accordingly, identifying mechanisms that activate DNMT3A is of both fundamental and therapeutic interest. Here, we applied a base editor mutational scanning strategy with an improved DNA methylation reporter to systematically identify DNMT3A activating mutations in cells. By integrating an optimized cellular recruitment strategy with paired isogenic cell lines with or without the LOF hotspot R882H mutation, we identify and validate three distinct hyperactivating mutations within or interacting with the regulatory ADD domain of DNMT3A, nominating these regions as potential functional target sites for pharmacological intervention. Notably, these mutations are still activating in the context of a heterozygous R882H mutation. Altogether, we showcase the utility of base editor scanning for discovering functional regions of target proteins.
Conflict of interest statement
B.B.L. holds sponsored research projects with AstraZeneca and Eisai, is a scientific consultant for Imago BioSciences and Exo Therapeutics and is a shareholder and member of the scientific advisory board of Light Horse Therapeutics.
Figures



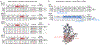
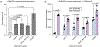
References
-
- Russler-Germain DA; Spencer DH; Young MA; Lamprecht TL; Miller CA; Fulton R; Meyer MR; Erdmann-Gilmore P; Townsend RR; Wilson RK; Ley TJ The R882H DNMT3A Mutation Associated with AML Dominantly Inhibits Wild-Type DNMT3A by Blocking Its Ability to Form Active Tetramers. Cancer Cell 2014, 25 (4), 442–454. 10.1016/j.ccr.2014.02.010. - DOI - PMC - PubMed
-
- Nguyen T-V; Yao S; Wang Y; Rolfe A; Selvaraj A; Darman R; Ke J; Warmuth M; Smith PG; Larsen NA; Yu L; Zhu P; Fekkes P; Vaillancourt FH; Bolduc DM The R882H DNMT3A Hot Spot Mutation Stabilizes the Formation of Large DNMT3A Oligomers with Low DNA Methyltransferase Activity. J. Biol. Chem 2019, 294 (45), 16966–16977. 10.1074/jbc.RA119.010126. - DOI - PMC - PubMed
-
- Huang Y-H; Chen C-W; Sundaramurthy V; Słabicki M; Hao D; Watson CJ; Tovy A; Reyes JM; Dakhova O; Crovetti BR; Galonska C; Lee M; Brunetti L; Zhou Y; Tatton-Brown K; Huang Y; Cheng X; Meissner A; Valk PJM; Van Maldergem L; Sanders MA; Blundell JR; Li W; Ebert BL; Goodell MA Systematic Profiling of DNMT3A Variants Reveals Protein Instability Mediated by the DCAF8 E3 Ubiquitin Ligase Adaptor. Cancer Discov. 2022, 12 (1), 220–235. 10.1158/2159-8290.CD-21-0560. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

