Allotopic expression of COX6 elucidates Atco-driven co-assembly of cytochrome oxidase and ATP synthase
- PMID: 37604582
- PMCID: PMC10442929
- DOI: 10.26508/lsa.202301965
Allotopic expression of COX6 elucidates Atco-driven co-assembly of cytochrome oxidase and ATP synthase
Abstract
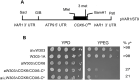
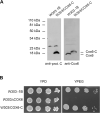
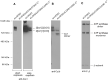
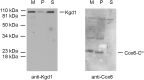
The Cox6 subunit of Saccharomyces cerevisiae cytochrome oxidase (COX) and the Atp9 subunit of the ATP synthase are encoded in nuclear and mitochondrial DNA, respectively. The two proteins interact to form Atco complexes that serve as the source of Atp9 for ATP synthase assembly. To determine if Atco is also a precursor of COX, we pulse-labeled Cox6 in isolated mitochondria of a cox6 nuclear mutant with COX6 in mitochondrial DNA. Only a small fraction of the newly translated Cox6 was found to be present in Atco, which can explain the low concentration of COX and poor complementation of the cox6 mutation by the allotopic gene. This and other pieces of evidence presented in this study indicate that Atco is an obligatory source of Cox6 for COX biogenesis. Together with our finding that atp9 mutants fail to assemble COX, we propose a regulatory model in which Atco unidirectionally couples the biogenesis of COX to that of the ATP synthase to maintain a proper ratio of these two complexes of oxidative phosphorylation.
© 2023 Veloso R Franco et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







References
-
- Böttinger L, Guiard B, Oeljeklaus S, Kulawiak B, Zufall N, Wiedemann N, Warscheid B, van der Laan M, Becker T (2013) A complex of Cox4 and mitochondrial Hsp70 plays an important role in the assembly of the cytochrome c oxidase. Mol Biol Cell 24: 2609–2619. 10.1091/mbc.E13-02-0106 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
