The Impact of Chromosomal Rearrangements in Speciation: From Micro- to Macroevolution
- PMID: 37604585
- PMCID: PMC10626258
- DOI: 10.1101/cshperspect.a041447
The Impact of Chromosomal Rearrangements in Speciation: From Micro- to Macroevolution
Abstract
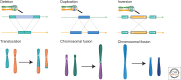
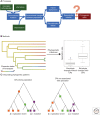
Chromosomal rearrangements (CRs) have been known since almost the beginning of genetics. While an important role for CRs in speciation has been suggested, evidence primarily stems from theoretical and empirical studies focusing on the microevolutionary level (i.e., on taxon pairs where speciation is often incomplete). Although the role of CRs in eukaryotic speciation at a macroevolutionary level has been supported by associations between species diversity and rates of evolution of CRs across phylogenies, these findings are limited to a restricted range of CRs and taxa. Now that more broadly applicable and precise CR detection approaches have become available, we address the challenges in filling some of the conceptual and empirical gaps between micro- and macroevolutionary studies on the role of CRs in speciation. We synthesize what is known about the macroevolutionary impact of CRs and suggest new research avenues to overcome the pitfalls of previous studies to gain a more comprehensive understanding of the evolutionary significance of CRs in speciation across the tree of life.
Copyright © 2023 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Ahola V, Lehtonen R, Somervuo P, Salmela L, Koskinen P, Rastas P, Välimäki N, Paulin L, Kvist J, Wahlberg N, et al. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun 5: 4737. 10.1038/ncomms5737 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
