GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data
- PMID: 37604963
- PMCID: PMC10904014
- DOI: 10.1038/s41588-023-01449-0
GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data
Erratum in
-
Author Correction: GATK-gCNV enables the discovery of rare copy number variants from exome sequencing data.Nat Genet. 2024 Mar;56(3):553. doi: 10.1038/s41588-024-01663-4. Nat Genet. 2024. PMID: 38263447 No abstract available.
Abstract
Copy number variants (CNVs) are major contributors to genetic diversity and disease. While standardized methods, such as the genome analysis toolkit (GATK), exist for detecting short variants, technical challenges have confounded uniform large-scale CNV analyses from whole-exome sequencing (WES) data. Given the profound impact of rare and de novo coding CNVs on genome organization and human disease, we developed GATK-gCNV, a flexible algorithm to discover rare CNVs from sequencing read-depth information, complete with open-source distribution via GATK. We benchmarked GATK-gCNV in 7,962 exomes from individuals in quartet families with matched genome sequencing and microarray data, finding up to 95% recall of rare coding CNVs at a resolution of more than two exons. We used GATK-gCNV to generate a reference catalog of rare coding CNVs in WES data from 197,306 individuals in the UK Biobank, and observed strong correlations between per-gene CNV rates and measures of mutational constraint, as well as rare CNV associations with multiple traits. In summary, GATK-gCNV is a tunable approach for sensitive and specific CNV discovery in WES data, with broad applications.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures
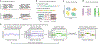


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

