Peptide foldamer-based inhibitors of the SARS-CoV-2 S protein-human ACE2 interaction
- PMID: 37605435
- PMCID: PMC10446788
- DOI: 10.1080/14756366.2023.2244693
Peptide foldamer-based inhibitors of the SARS-CoV-2 S protein-human ACE2 interaction
Abstract
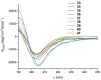
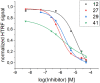
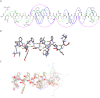
The entry of the SARS-CoV-2 virus into a human host cell begins with the interaction between the viral spike protein (S protein) and human angiotensin-converting enzyme 2 (hACE2). Therefore, a possible strategy for the treatment of this infection is based on inhibiting the interaction of the two abovementioned proteins. Compounds that bind to the SARS-CoV-2 S protein at the interface with the alpha-1/alpha-2 helices of ACE2 PD Subdomain I are of particular interest. We present a stepwise optimisation of helical peptide foldamers containing trans-2-aminocylopentanecarboxylic acid residues as the folding-inducing unit. Four rounds of optimisation led to the discovery of an 18-amino-acid peptide with high affinity for the SARS-CoV-2 S protein (Kd = 650 nM) that inhibits this protein-protein interaction with IC50 = 1.3 µM. Circular dichroism and nuclear magnetic resonance studies indicated the helical conformation of this peptide in solution.
Keywords: BLI; COVID-19; foldamers; helix; peptides; protein-protein interaction.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures









Similar articles
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection.Commun Biol. 2021 Feb 12;4(1):197. doi: 10.1038/s42003-021-01736-8. Commun Biol. 2021. PMID: 33580154 Free PMC article.
-
Label-Free Analysis of Binding and Inhibition of SARS-Cov-19 Spike Proteins to ACE2 Receptor with ACE2-Derived Peptides by Surface Plasmon Resonance.ACS Appl Bio Mater. 2023 Jan 16;6(1):182-190. doi: 10.1021/acsabm.2c00832. Epub 2022 Dec 22. ACS Appl Bio Mater. 2023. PMID: 36550079 Free PMC article.
-
Engineering defensin α-helix to produce high-affinity SARS-CoV-2 spike protein binding ligands.Protein Sci. 2022 Jun;31(6):e4355. doi: 10.1002/pro.4355. Protein Sci. 2022. PMID: 35634778 Free PMC article.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
Cited by
-
Scalable Synthesis of All Stereoisomers of 2-Aminocyclopentanecarboxylic Acid─A Toolbox for Peptide Foldamer Chemistry.J Org Chem. 2024 Apr 5;89(7):4760-4767. doi: 10.1021/acs.joc.3c02991. Epub 2024 Mar 27. J Org Chem. 2024. PMID: 38544408 Free PMC article.
-
Antiviral Protein-Protein Interaction Inhibitors.J Med Chem. 2024 Mar 14;67(5):3205-3231. doi: 10.1021/acs.jmedchem.3c01543. Epub 2024 Feb 23. J Med Chem. 2024. PMID: 38394369 Free PMC article. Review.
-
Photoresponsive Helical Foldamers: Conformational Control Through Double Helix Formation and Light-Induced Protonation.Chemistry. 2025 Feb 20;31(11):e202403771. doi: 10.1002/chem.202403771. Epub 2025 Jan 16. Chemistry. 2025. PMID: 39749723 Free PMC article.
-
Inhibition of ACE2-S Protein Interaction by a Short Functional Peptide with a Boomerang Structure.Molecules. 2024 Jun 26;29(13):3022. doi: 10.3390/molecules29133022. Molecules. 2024. PMID: 38998974 Free PMC article.
References
-
- Cheng RP, Gellman SH, DeGrado WF.. β-peptides: from structure to function. Chem Rev. 2001;101(10):3219–3232. - PubMed
-
- Gellman SH. Foldamers: a manifesto. Acc Chem Res. 1998;31(4):173–180.
-
- Martinek TA, Fülöp F.. Peptidic foldamers: ramping up diversity. Chem. Soc. Rev. 2012;41(2):687–702. - PubMed
-
- Seebach D, Overhand M, Kühnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. β Peptides: synthesis by Arndt-Eistert homologation with concomitant peptide coupling. Structure determination by NMR and CD spectroscopy and by X-ray crystallography. Helical secondary structure of a β-hexapeptide in solution and its stability towards Pe. HCA. 1996;79(4):913–941.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
