Translation-coupled mRNA quality control mechanisms
- PMID: 37605642
- PMCID: PMC10548175
- DOI: 10.15252/embj.2023114378
Translation-coupled mRNA quality control mechanisms
Abstract
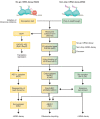
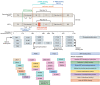
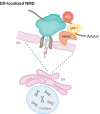
mRNA surveillance pathways are essential for accurate gene expression and to maintain translation homeostasis, ensuring the production of fully functional proteins. Future insights into mRNA quality control pathways will enable us to understand how cellular mRNA levels are controlled, how defective or unwanted mRNAs can be eliminated, and how dysregulation of these can contribute to human disease. Here we review translation-coupled mRNA quality control mechanisms, including the non-stop and no-go mRNA decay pathways, describing their mechanisms, shared trans-acting factors, and differences. We also describe advances in our understanding of the nonsense-mediated mRNA decay (NMD) pathway, highlighting recent mechanistic findings, the discovery of novel factors, as well as the role of NMD in cellular physiology and its impact on human disease.
Keywords: No-go mRNA decay; Non-stop mRNA decay; Nonsense-mediated mRNA decay; RNA quality control; UPF1.
© 2023 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
Javier F Caceres is a member of the Advisory Editorial Board of The EMBO Journal. This has no bearing on the editorial consideration of this article for publication.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

