Review of knockout technology approaches in bacterial drug resistance research
- PMID: 37605748
- PMCID: PMC10440060
- DOI: 10.7717/peerj.15790
Review of knockout technology approaches in bacterial drug resistance research
Abstract
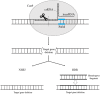
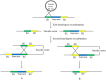
Gene knockout is a widely used method in biology for investigating gene function. Several technologies are available for gene knockout, including zinc-finger nuclease technology (ZFN), suicide plasmid vector systems, transcription activator-like effector protein nuclease technology (TALEN), Red homologous recombination technology, CRISPR/Cas, and others. Of these, Red homologous recombination technology, CRISPR/Cas9 technology, and suicide plasmid vector systems have been the most extensively used for knocking out bacterial drug resistance genes. These three technologies have been shown to yield significant results in researching bacterial gene functions in numerous studies. This study provides an overview of current gene knockout methods that are effective for genetic drug resistance testing in bacteria. The study aims to serve as a reference for selecting appropriate techniques.
Keywords: Bacterial drug resistance; CRISPR/Cas9; Gene knockout; Red homologous recombination; Suicide plasmid.
© 2023 Tong et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Genome Editing of Silkworms.Methods Mol Biol. 2023;2637:359-374. doi: 10.1007/978-1-0716-3016-7_27. Methods Mol Biol. 2023. PMID: 36773160
-
Basics of genome editing technology and its application in livestock species.Reprod Domest Anim. 2017 Aug;52 Suppl 3:4-13. doi: 10.1111/rda.13012. Reprod Domest Anim. 2017. PMID: 28815851 Review.
-
Genome Editing of Rat.Methods Mol Biol. 2023;2637:223-231. doi: 10.1007/978-1-0716-3016-7_17. Methods Mol Biol. 2023. PMID: 36773150
-
Application and development of genome editing technologies to the Solanaceae plants.Plant Physiol Biochem. 2018 Oct;131:37-46. doi: 10.1016/j.plaphy.2018.02.019. Epub 2018 Mar 2. Plant Physiol Biochem. 2018. PMID: 29523384 Review.
-
Genome Editing with mRNA Encoding ZFN, TALEN, and Cas9.Mol Ther. 2019 Apr 10;27(4):735-746. doi: 10.1016/j.ymthe.2019.01.014. Epub 2019 Jan 25. Mol Ther. 2019. PMID: 30803822 Free PMC article. Review.
Cited by
-
Genomic engineering in Rhizobium etli: implementation and evaluation of systems based on dCas9.Front Microbiol. 2025 Jun 24;16:1604430. doi: 10.3389/fmicb.2025.1604430. eCollection 2025. Front Microbiol. 2025. PMID: 40630179 Free PMC article.
-
Carvacrol and Streptomycin in Combination Weaken Streptomycin Resistance in Pectobacterium carotovorum subsp. carotovorum.Plants (Basel). 2025 Mar 14;14(6):908. doi: 10.3390/plants14060908. Plants (Basel). 2025. PMID: 40265826 Free PMC article.
-
Mandelonitrile produced by commensal bacteria protects the Colorado potato beetle against predation.Nat Commun. 2024 Nov 21;15(1):10081. doi: 10.1038/s41467-024-54439-z. Nat Commun. 2024. PMID: 39572567 Free PMC article.
-
A Simple Allelic Exchange Method for Efficient Seamless Knockout of Up to 34-kbp-Long Gene Cassettes in Pseudomonas.Appl Biochem Biotechnol. 2024 Aug;196(8):5616-5630. doi: 10.1007/s12010-023-04806-1. Epub 2023 Dec 16. Appl Biochem Biotechnol. 2024. PMID: 38103122
-
Advancements in gene editing technologies for probiotic-enabled disease therapy.iScience. 2024 Aug 22;27(9):110791. doi: 10.1016/j.isci.2024.110791. eCollection 2024 Sep 20. iScience. 2024. PMID: 39286511 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

