Network analysis identifies strain-dependent response to tau and tau seeding-associated genes
- PMID: 37606887
- PMCID: PMC10443211
- DOI: 10.1084/jem.20230180
Network analysis identifies strain-dependent response to tau and tau seeding-associated genes
Abstract
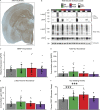
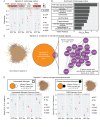
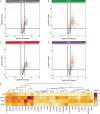
Previous research demonstrated that genetic heterogeneity is a critical factor in modeling amyloid accumulation and other Alzheimer's disease phenotypes. However, it is unknown what mechanisms underlie these effects of genetic background on modeling tau aggregate-driven pathogenicity. In this study, we induced tau aggregation in wild-derived mice by expressing MAPT. To investigate the effect of genetic background on the action of tau aggregates, we performed RNA sequencing with brains of C57BL/6J, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice (n = 64) and determined core transcriptional signature conserved in all genetic backgrounds and signature unique to wild-derived backgrounds. By measuring tau seeding activity using the cortex, we identified 19 key genes associated with tau seeding and amyloid response. Interestingly, microglial pathways were strongly associated with tau seeding activity in CAST/EiJ and PWK/PhJ backgrounds. Collectively, our study demonstrates that mouse genetic context affects tau-mediated alteration of transcriptome and tau seeding. The gene modules associated with tau seeding provide an important resource to better model tauopathy.
© 2023 Acri et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures












Update of
-
Network analysis reveals strain-dependent response to misfolded tau aggregates.bioRxiv [Preprint]. 2023 Jan 30:2023.01.28.526029. doi: 10.1101/2023.01.28.526029. bioRxiv. 2023. Update in: J Exp Med. 2023 Nov 6;220(11):e20230180. doi: 10.1084/jem.20230180. PMID: 36778440 Free PMC article. Updated. Preprint.
References
-
- Bengoa-Vergniory, N., Velentza-Almpani E., Silva A.M., Scott C., Vargas-Caballero M., Sastre M., Wade-Martins R., and Alegre-Abarrategui J.. 2021. Tau-proximity ligation assay reveals extensive previously undetected pathology prior to neurofibrillary tangles in preclinical Alzheimer’s disease. Acta Neuropathol. Commun. 9:18. 10.1186/s40478-020-01117-y - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- R01 AG077829/AG/NIA NIH HHS/United States
- T32 AG071444/AG/NIA NIH HHS/United States
- R21 AG072738/AG/NIA NIH HHS/United States
- F31 AG074673/AG/NIA NIH HHS/United States
- T32 DK064466/DK/NIDDK NIH HHS/United States
- RF1 AG062077/AG/NIA NIH HHS/United States
- RF1 AG074543/AG/NIA NIH HHS/United States
- F30 AG079580/AG/NIA NIH HHS/United States
- R01 AG071281/AG/NIA NIH HHS/United States
- T32 GM077229/GM/NIGMS NIH HHS/United States
- T32 GM148382/GM/NIGMS NIH HHS/United States
- F31 AG074628/AG/NIA NIH HHS/United States
- R01 NS119280/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

