Deciphering Protein Secretion from the Brain to Cerebrospinal Fluid for Biomarker Discovery
- PMID: 37606934
- PMCID: PMC10476268
- DOI: 10.1021/acs.jproteome.3c00366
Deciphering Protein Secretion from the Brain to Cerebrospinal Fluid for Biomarker Discovery
Abstract
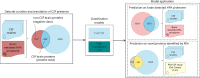
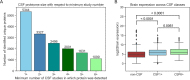
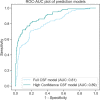
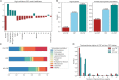
Cerebrospinal fluid (CSF) is an essential matrix for the discovery of neurological disease biomarkers. However, the high dynamic range of protein concentrations in CSF hinders the detection of the least abundant protein biomarkers by untargeted mass spectrometry. It is thus beneficial to gain a deeper understanding of the secretion processes within the brain. Here, we aim to explore if and how the secretion of brain proteins to the CSF can be predicted. By combining a curated CSF proteome and the brain elevated proteome of the Human Protein Atlas, brain proteins were classified as CSF or non-CSF secreted. A machine learning model was trained on a range of sequence-based features to differentiate between CSF and non-CSF groups and effectively predict the brain origin of proteins. The classification model achieves an area under the curve of 0.89 if using high confidence CSF proteins. The most important prediction features include the subcellular localization, signal peptides, and transmembrane regions. The classifier generalized well to the larger brain detected proteome and is able to correctly predict novel CSF proteins identified by affinity proteomics. In addition to elucidating the underlying mechanisms of protein secretion, the trained classification model can support biomarker candidate selection.
Keywords: brain proteome; cerebrospinal fluid; fluid biomarker; machine learning; protein secretion.
Conflict of interest statement
The authors declare the following competing financial interest(s): C.E.T. has a collaboration contract with ADx Neurosciences, Quanterix and Eli Lilly, performed contract research or received grants from AC-Immune, Axon Neurosciences, Biogen, Brainstorm Therapeutics, Celgene, EIP Pharma, Eisai, PeopleBio Inc., Roche, Toyama, Vivoryon, and has a speaker contract with Roche. S.A. reports grants and nonfinancial support from Cergentis BV and a patent pending outside the submitted work. The MIRIADE project includes the following commercial beneficiaries and partners: ADx Neuroscience, ENPICOM, LGC Limited, PeopleBio Inc., Olink, Quanterix, and Roche.
Figures


References
-
- Teunissen C. E.; Otto M.; Engelborghs S.; Herukka S.-K.; Lehmann S.; Lewczuk P.; Lleó A.; Perret-Liaudet A.; Tumani H.; Turner M. R.; Verbeek M. M.; Wiltfang J.; Zetterberg H.; Parnetti L.; Blennow K. White Paper by the Society for CSF Analysis and Clinical Neurochemistry: Overcoming Barriers in Biomarker Development and Clinical Translation. Alz Res. Therapy 2018, 10 (1), 30. 10.1186/s13195-018-0359-x. - DOI - PMC - PubMed
-
- Kroksveen A. C.; Opsahl J. A.; Aye T. T.; Ulvik R. J.; Berven F. S. Proteomics of Human Cerebrospinal Fluid: Discovery and Verification of Biomarker Candidates in Neurodegenerative Diseases Using Quantitative Proteomics. Journal of Proteomics 2011, 74 (4), 371–388. 10.1016/j.jprot.2010.11.010. - DOI - PubMed
-
- Duits F. H.; Martinez-Lage P.; Paquet C.; Engelborghs S.; Lleó A.; Hausner L.; Molinuevo J. L.; Stomrud E.; Farotti L.; Ramakers I. H. G. B.; Tsolaki M.; Skarsgård C.; Åstrand R.; Wallin A.; Vyhnalek M.; Holmber-Clausen M.; Forlenza O. V.; Ghezzi L.; Ingelsson M.; Hoff E. I.; Roks G.; Mendonça A.; Papma J. M.; Izagirre A.; Taga M.; Struyfs H.; Alcolea D. A.; Frölich L.; Balasa M.; Minthon L.; Twisk J. W. R.; Persson S.; Zetterberg H.; Flier W. M.; Teunissen C. E.; Scheltens P.; Blennow K. Performance and Complications of Lumbar Puncture in Memory Clinics: Results of the Multicenter Lumbar Puncture Feasibility Study. Alzheimer’s & Dementia 2016, 12 (2), 154–163. 10.1016/j.jalz.2015.08.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

