Somatic rearrangements causing oncogenic ectodomain deletions of FGFR1 in squamous cell lung cancer
- PMID: 37606995
- PMCID: PMC10617767
- DOI: 10.1172/JCI170217
Somatic rearrangements causing oncogenic ectodomain deletions of FGFR1 in squamous cell lung cancer
Abstract
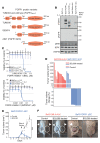
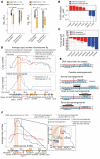
The discovery of frequent 8p11-p12 amplifications in squamous cell lung cancer (SQLC) has fueled hopes that FGFR1, located inside this amplicon, might be a therapeutic target. In a clinical trial, only 11% of patients with 8p11 amplification (detected by FISH) responded to FGFR kinase inhibitor treatment. To understand the mechanism of FGFR1 dependency, we performed deep genomic characterization of 52 SQLCs with 8p11-p12 amplification, including 10 tumors obtained from patients who had been treated with FGFR inhibitors. We discovered somatically altered variants of FGFR1 with deletion of exons 1-8 that resulted from intragenic tail-to-tail rearrangements. These ectodomain-deficient FGFR1 variants (ΔEC-FGFR1) were expressed in the affected tumors and were tumorigenic in both in vitro and in vivo models of lung cancer. Mechanistically, breakage-fusion-bridges were the source of 8p11-p12 amplification, resulting from frequent head-to-head and tail-to-tail rearrangements. Generally, tail-to-tail rearrangements within or in close proximity upstream of FGFR1 were associated with FGFR1 dependency. Thus, the genomic events shaping the architecture of the 8p11-p12 amplicon provide a mechanistic explanation for the emergence of FGFR1-driven SQLC. Specifically, we believe that FGFR1 ectodomain-deficient and FGFR1-centered amplifications caused by tail-to-tail rearrangements are a novel somatic genomic event that might be predictive of therapeutically relevant FGFR1 dependency.
Keywords: Drug therapy; Genetics; Lung cancer; Oncology.
Conflict of interest statement
Figures



Comment in
-
Genomic insights into the mechanisms of FGFR1 dependency in squamous cell lung cancer.J Clin Invest. 2023 Nov 1;133(21):e174171. doi: 10.1172/JCI174171. J Clin Invest. 2023. PMID: 37909331 Free PMC article.
-
High-resolution genomic configuration of FGFR rearrangements dictates the therapeutic vulnerability of squamous cell lung cancers.Transl Lung Cancer Res. 2024 Feb 29;13(2):236-239. doi: 10.21037/tlcr-23-705. Epub 2024 Feb 20. Transl Lung Cancer Res. 2024. PMID: 38496689 Free PMC article. No abstract available.
-
Oncogene alterations in non-small cell lung cancer with FGFR1 amplification-novel approach to stratify patients who benefit from FGFR inhibitors.Transl Lung Cancer Res. 2024 Mar 29;13(3):684-688. doi: 10.21037/tlcr-23-777. Epub 2024 Mar 8. Transl Lung Cancer Res. 2024. PMID: 38601453 Free PMC article. No abstract available.
References
-
- Nogova L, et al. Evaluation of BGJ398, a fibroblast growth factor receptor 1-3 kinaseinhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global phase I, dose-escalation and dose-expansion study. J Clin Oncol. 2017;35(2):157–165. doi: 10.1200/JCO.2016.67.2048. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

