Phenotype screens of murine pancreatic cancer identify a Tgf-α-Ccl2-paxillin axis driving human-like neural invasion
- PMID: 37607005
- PMCID: PMC10617783
- DOI: 10.1172/JCI166333
Phenotype screens of murine pancreatic cancer identify a Tgf-α-Ccl2-paxillin axis driving human-like neural invasion
Abstract
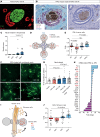
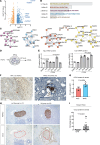
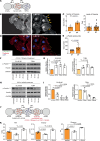
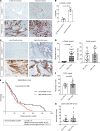
Solid cancers like pancreatic ductal adenocarcinoma (PDAC), a type of pancreatic cancer, frequently exploit nerves for rapid dissemination. This neural invasion (NI) is an independent prognostic factor in PDAC, but insufficiently modeled in genetically engineered mouse models (GEMM) of PDAC. Here, we systematically screened for human-like NI in Europe's largest repository of GEMM of PDAC, comprising 295 different genotypes. This phenotype screen uncovered 2 GEMMs of PDAC with human-like NI, which are both characterized by pancreas-specific overexpression of transforming growth factor α (TGF-α) and conditional depletion of p53. Mechanistically, cancer-cell-derived TGF-α upregulated CCL2 secretion from sensory neurons, which induced hyperphosphorylation of the cytoskeletal protein paxillin via CCR4 on cancer cells. This activated the cancer migration machinery and filopodia formation toward neurons. Disrupting CCR4 or paxillin activity limited NI and dampened tumor size and tumor innervation. In human PDAC, phospho-paxillin and TGF-α-expression constituted strong prognostic factors. Therefore, we believe that the TGF-α-CCL2-CCR4-p-paxillin axis is a clinically actionable target for constraining NI and tumor progression in PDAC.
Keywords: Cancer; Innervation; Oncology.
Figures
Similar articles
-
Kindlin-2 induced by TGF-β signaling promotes pancreatic ductal adenocarcinoma progression through downregulation of transcriptional factor HOXB9.Cancer Lett. 2015 May 28;361(1):75-85. doi: 10.1016/j.canlet.2015.02.039. Epub 2015 Feb 24. Cancer Lett. 2015. PMID: 25724625
-
CXCL10 and CCL21 Promote Migration of Pancreatic Cancer Cells Toward Sensory Neurons and Neural Remodeling in Tumors in Mice, Associated With Pain in Patients.Gastroenterology. 2020 Aug;159(2):665-681.e13. doi: 10.1053/j.gastro.2020.04.037. Epub 2020 Apr 21. Gastroenterology. 2020. PMID: 32330476
-
Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway.J Exp Clin Cancer Res. 2019 Jul 15;38(1):310. doi: 10.1186/s13046-019-1313-x. J Exp Clin Cancer Res. 2019. PMID: 31307515 Free PMC article.
-
FAK and paxillin, two potential targets in pancreatic cancer.Oncotarget. 2016 May 24;7(21):31586-601. doi: 10.18632/oncotarget.8040. Oncotarget. 2016. PMID: 26980710 Free PMC article. Review.
-
Modeling pancreatic cancer in mice for experimental therapeutics.Biochim Biophys Acta Rev Cancer. 2021 Aug;1876(1):188554. doi: 10.1016/j.bbcan.2021.188554. Epub 2021 May 1. Biochim Biophys Acta Rev Cancer. 2021. PMID: 33945847 Free PMC article. Review.
Cited by
-
Placental growth factor promotes neural invasion and predicts disease prognosis in resectable pancreatic cancer.J Exp Clin Cancer Res. 2024 May 30;43(1):153. doi: 10.1186/s13046-024-03066-z. J Exp Clin Cancer Res. 2024. PMID: 38816706 Free PMC article.
-
Anti-metastatic potential of flavonoids for the treatment of cancers: focus on epithelial-mesenchymal transition (EMT) process.Naunyn Schmiedebergs Arch Pharmacol. 2025 May 28. doi: 10.1007/s00210-025-04235-3. Online ahead of print. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 40434422 Review.
-
Pinpointing neurons that hinder cancer treatment.Nature. 2025 Apr;640(8060):889-890. doi: 10.1038/d41586-025-00501-9. Nature. 2025. PMID: 39966586 No abstract available.
-
Tumor-infiltrating nerves: unraveling the role of cancer neuroscience in tumorigenesis, disease progression, and emerging therapies.Discov Oncol. 2025 Jul 1;16(1):1209. doi: 10.1007/s12672-025-02827-2. Discov Oncol. 2025. PMID: 40591169 Free PMC article. Review.
-
Targeting the NPY/NPY1R signaling axis in mutant p53-dependent pancreatic cancer impairs metastasis.Sci Adv. 2025 Mar 14;11(11):eadq4416. doi: 10.1126/sciadv.adq4416. Epub 2025 Mar 12. Sci Adv. 2025. PMID: 40073121 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

