Characterization of a mouse-adapted strain of bat severe acute respiratory syndrome-related coronavirus
- PMID: 37607058
- PMCID: PMC10537601
- DOI: 10.1128/jvi.00790-23
Characterization of a mouse-adapted strain of bat severe acute respiratory syndrome-related coronavirus
Abstract
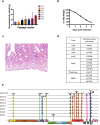
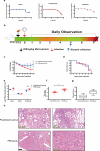
Bats carry genetically diverse severe acute respiratory syndrome-related coronaviruses (SARSr-CoVs). Some of them utilize human angiotensin-converting enzyme 2 (hACE2) as a receptor and cannot efficiently replicate in wild-type mice. Our previous study demonstrated that the bat SARSr-CoV rRsSHC014S induces respiratory infection and lung damage in hACE2 transgenic mice but not wild-type mice. In this study, we generated a mouse-adapted strain of rRsSHC014S, which we named SMA1901, by serial passaging of wild-type virus in BALB/c mice. SMA1901 showed increased infectivity in mouse lungs and induced interstitial lung pneumonia in both young and aged mice after intranasal inoculation. Genome sequencing revealed mutations in not only the spike protein but the whole genome, which may be responsible for the enhanced pathogenicity of SMA1901 in wild-type BALB/c mice. SMA1901 induced age-related mortality similar to that observed in SARS and COVID-19. Drug testing using antibodies and antiviral molecules indicated that this mouse-adapted virus strain can be used to test prophylactic and therapeutic drug candidates against SARSr-CoVs. IMPORTANCE The genetic diversity of SARSr-CoVs in wildlife and their potential risk of cross-species infection highlights the importance of developing a powerful animal model to evaluate the antibodies and antiviral drugs. We acquired the mouse-adapted strain of a bat-origin coronavirus named SMA1901 by natural serial passaging of rRsSHC014S in BALB/c mice. The SMA1901 infection caused interstitial pneumonia and inflammatory immune responses in both young and aged BALB/c mice after intranasal inoculation. Our model exhibited age-related mortality similar to SARS and COVID-19. Therefore, our model will be of high value for investigating the pathogenesis of bat SARSr-CoVs and could serve as a prospective test platform for prophylactic and therapeutic candidates.
Keywords: animal model; bat SARS-related coronavirus; mouse-adapted strain; pathogenicity; pneumonia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Inactivated SARS-CoV-2 Vaccine Shows Cross-Protection against Bat SARS-Related Coronaviruses in Human ACE2 Transgenic Mice.J Virol. 2022 Apr 27;96(8):e0016922. doi: 10.1128/jvi.00169-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343762 Free PMC article.
-
Evolutionary Arms Race between Virus and Host Drives Genetic Diversity in Bat Severe Acute Respiratory Syndrome-Related Coronavirus Spike Genes.J Virol. 2020 Sep 29;94(20):e00902-20. doi: 10.1128/JVI.00902-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32699095 Free PMC article.
-
Epidemiology and Genomic Characterization of Two Novel SARS-Related Coronaviruses in Horseshoe Bats from Guangdong, China.mBio. 2022 Jun 28;13(3):e0046322. doi: 10.1128/mbio.00463-22. Epub 2022 Apr 25. mBio. 2022. PMID: 35467426 Free PMC article.
-
Molecular epidemiology, evolution and phylogeny of SARS coronavirus.Infect Genet Evol. 2019 Jul;71:21-30. doi: 10.1016/j.meegid.2019.03.001. Epub 2019 Mar 4. Infect Genet Evol. 2019. PMID: 30844511 Free PMC article. Review.
-
Geographical structure of bat SARS-related coronaviruses.Infect Genet Evol. 2019 Apr;69:224-229. doi: 10.1016/j.meegid.2019.02.001. Epub 2019 Feb 6. Infect Genet Evol. 2019. PMID: 30735813 Free PMC article. Review.
Cited by
-
Characterization of a SARS-CoV-2 infection model in golden hamsters with diabetes mellitus.Virol Sin. 2025 Jun;40(3):349-360. doi: 10.1016/j.virs.2025.05.001. Epub 2025 May 17. Virol Sin. 2025. PMID: 40389095 Free PMC article.
-
UB-612 pan-SARS-CoV-2 T cell immunity-promoting vaccine protects against COVID-19 moderate-severe disease.iScience. 2024 Jan 17;27(2):108887. doi: 10.1016/j.isci.2024.108887. eCollection 2024 Feb 16. iScience. 2024. PMID: 38318376 Free PMC article.
-
Broad cross neutralizing antibodies against sarbecoviruses generated by SARS-CoV-2 infection and vaccination in humans.NPJ Vaccines. 2024 Oct 22;9(1):195. doi: 10.1038/s41541-024-00997-8. NPJ Vaccines. 2024. PMID: 39438493 Free PMC article.
References
-
- Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM, Berger A, Burguière A-M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J-C, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk H-D, Osterhaus ADME, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi:10.1056/NEJMoa030747 - DOI - PubMed
-
- Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588:E6. doi:10.1038/s41586-020-2951-z - DOI - PMC - PubMed
-
- Yang XL, Hu B, Wang B, Wang MN, Zhang Q, Zhang W, Wu LJ, Ge XY, Zhang YZ, Daszak P, Wang LF, Shi ZL. 2015. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol 90:3253–3256. doi:10.1128/JVI.02582-15 - DOI - PMC - PubMed
-
- Hu B, Zeng L-P, Yang X-L, Ge X-Y, Zhang W, Li B, Xie J-Z, Shen X-R, Zhang Y-Z, Wang N, Luo D-S, Zheng X-S, Wang M-N, Daszak P, Wang L-F, Cui J, Shi Z-L, Drosten C. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 13:e1006698. doi:10.1371/journal.ppat.1006698 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

