Bcl6, Irf2, and Notch2 promote nonclassical monocyte development
- PMID: 37607223
- PMCID: PMC10469339
- DOI: 10.1073/pnas.2220853120
Bcl6, Irf2, and Notch2 promote nonclassical monocyte development
Abstract
Ly6Clo monocytes are a myeloid subset that specializes in the surveillance of vascular endothelium. Ly6Clo monocytes have been shown to derive from Ly6Chi monocytes. NOTCH2 signaling has been implicated as a trigger for Ly6Clo monocyte development, but the basis for this effect is unclear. Here, we examined the impact of NOTCH2 signaling of myeloid progenitors on the development of Ly6Clo monocytes in vitro. NOTCH2 signaling induced by delta-like ligand 1 (DLL1) efficiently induced the transition of Ly6Chi TREML4- monocytes into Ly6Clo TREML4+ monocytes. We further identified two additional transcriptional requirements for development of Ly6Clo monocytes. Deletion of BCL6 from myeloid progenitors abrogated development of Ly6Clo monocytes. IRF2 was also required for Ly6Clo monocyte development in a cell-intrinsic manner. DLL1-induced in vitro transition into Ly6Clo TREML4+ monocytes required IRF2 but unexpectedly could occur in the absence of NUR77 or BCL6. These results imply a transcriptional hierarchy for these factors in controlling Ly6Clo monocyte development.
Keywords: Bcl6; IRF2; Notch2; nonclassical monocytes.
Conflict of interest statement
The authors declare no competing interest.
Figures




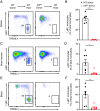
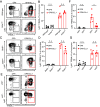
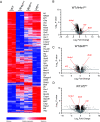
References
-
- Narasimhan P. B., Marcovecchio P., Hamers A. A. J., Hedrick C. C., Nonclassical monocytes in health and disease. Annu. Rev. Immunol. 37, 439–456 (2019). - PubMed
-
- Zembala M., Uracz W., Ruggiero I., Mytar B., Pryjma J., Isolation and functional characteristics of FcR+ and FcR- human monocyte subsets J. Immunol. 133, 1293–1299 (1984). - PubMed
-
- Passlick B., Flieger D., Ziegler-Heitbrock H. W., Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 74, 2527–2534 (1989). - PubMed
-
- Ziegler-Heitbrock H. W., et al. , The novel subset of CD14+/CD16+ blood monocytes exhibits features of tissue macrophages. Eur. J. Immunol. 23, 2053–2058 (1993). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

