Circadian modulation by time-restricted feeding rescues brain pathology and improves memory in mouse models of Alzheimer's disease
- PMID: 37607543
- PMCID: PMC10591997
- DOI: 10.1016/j.cmet.2023.07.014
Circadian modulation by time-restricted feeding rescues brain pathology and improves memory in mouse models of Alzheimer's disease
Abstract
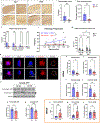
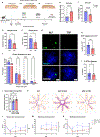
Circadian disruptions impact nearly all people with Alzheimer's disease (AD), emphasizing both their potential role in pathology and the critical need to investigate the therapeutic potential of circadian-modulating interventions. Here, we show that time-restricted feeding (TRF) without caloric restriction improved key disease components including behavioral timing, disease pathology, hippocampal transcription, and memory in two transgenic (TG) mouse models of AD. We found that TRF had the remarkable capability of simultaneously reducing amyloid deposition, increasing Aβ42 clearance, improving sleep and memory, and normalizing daily transcription patterns of multiple genes, including those associated with AD and neuroinflammation. Thus, our study unveils for the first time the pleiotropic nature of timed feeding on AD, which has far-reaching effects beyond metabolism, ameliorating neurodegeneration and the misalignment of circadian rhythmicity. Since TRF can substantially modify disease trajectory, this intervention has immediate translational potential, addressing the urgent demand for accessible approaches to reduce or halt AD progression.
Keywords: Alzheimer's mouse models; Alzheimer’s disease; amyloid plaque deposition; circadian rhythms; cognition; hippocampal transcriptome; memory; neuroinflammation; rhythmic transcription; time-restricted feeding.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Comment in
-
Restricting mealtime ameliorates neurodegeneration.Cell Metab. 2023 Oct 3;35(10):1673-1674. doi: 10.1016/j.cmet.2023.09.006. Cell Metab. 2023. PMID: 37793341
References
-
- Cronin P, McCarthy MJ, Lim ASP, Salmon DP, Galasko D, Masliah E, De Jager PL, Bennett DA, and Desplats P (2017). Circadian alterations during early stages of Alzheimer’s disease are associated with aberrant cycles of DNA methylation in BMAL1. Alzheimers Dement. 13, 689–700. 10.1016/j.jalz.2016.10.003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

